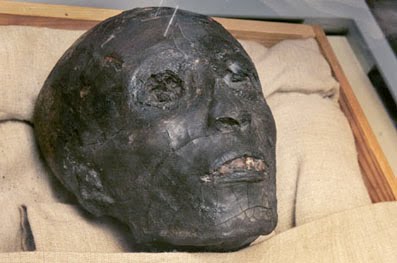
Despite the refusal of the Secretary General of the Egyptian Supreme Council of Antiquities, Zahi Hawass, to release any DNA results which might indicate the racial ancestry of Pharaoh Tutankhamen, the leaked results reveal that King Tut’s DNA is a 99.6 percent match with Western European Y chromosomes.
The DNA test results were inadvertently revealed on a Discovery Channel TV documentary filmed with Hawass’s permission — but it seems as if the Egyptian failed to spot the giveaway part of the documentary which revealed the test results.
Hawass previously announced that he would not release the racial DNA results of Egyptian mummies — obviously because he feared the consequences of such a revelation.
On the Discovery Channel broadcast, which can be seen on the Discovery Channel website here, or if they pull it, on YouTube here, at approximately 1:53 into the video, the camera pans over a printout of DNA test results from King Tut.
Firstly, here is a brief explanation of the results visible in the video. It is a list of what is called Short Tandem Repeats (STRs).
STRs are repeated DNA sequences which are “short repeat units” whose characteristics make them especially suitable for human identification.
These STR values for 17 markers visible in the video are as follows:
DYS 19 – 14 (? not clear)
DYS 385a – 11
DYS 385b – 14
DYS 389i – 13
DYS 389ii – 30
DYS 390 – 24
DYS 391 – 11
DYS 392 – 13
DYS 393 – 13
DYS 437 – 14 (? not clear)
DYS 438 – 12
DYS 439 – 10
DYS 448 – 19
DYS 456 – 15
DYS 458 – 16
DYS 635 – 23
YGATAH4 – 11
What does this mean? Fortunately, a genius by the name of Whit Athey provides the key to this list. Mr Athey is a retired physicist whose working career was primarily at the Food and Drug Administration where he was chief of one of the medical device labs.
Mr Athey received his doctorate in physics and biochemistry at Tufts University, and undergraduate (engineering) and masters (math) degrees at Auburn University. For several years during the 1980s, he also taught one course each semester in the electrical engineering department of the University of Maryland. Besides his interest in genetic genealogy, he is an amateur astronomer and has his own small observatory near his home in Brookeville, MD.
He also runs a very valuable website called the “Haplogroup Predictor” which allows users to input STR data and generate the haplogroup which marks those STR data.
For those who want to know what a haplogroup is, here is a “simple” definition: a haplogroup is a group of similar haplotypes that share a common ancestor with a single nucleotide polymorphism (SNP) mutation.
Still none the wiser? Damn these scientists.
Ok, let’s try it this way: a haplotype is a combination of multiple specific locations of a gene or DNA sequence on a chromosome.
Haplogroups are assigned letters of the alphabet, and refinements consist of additional number and letter combinations, for example R1b or R1b1. Y-chromosome and mitochondrial DNA haplogroups have different haplogroup designations. In essence, haplogroups give an inisight into ancestral origins dating back thousands of years.
By entering all the STR data inadvertently shown on the Discovery video, a 99.6 percent fit with the R1b haplogroup is revealed.
The significance is, of course, that R1b is the most common Y-chromosome haplogroup in Europe reaching its highest concentrations in Ireland, Scotland, western England and the European Atlantic seaboard — in other words, European through and through.
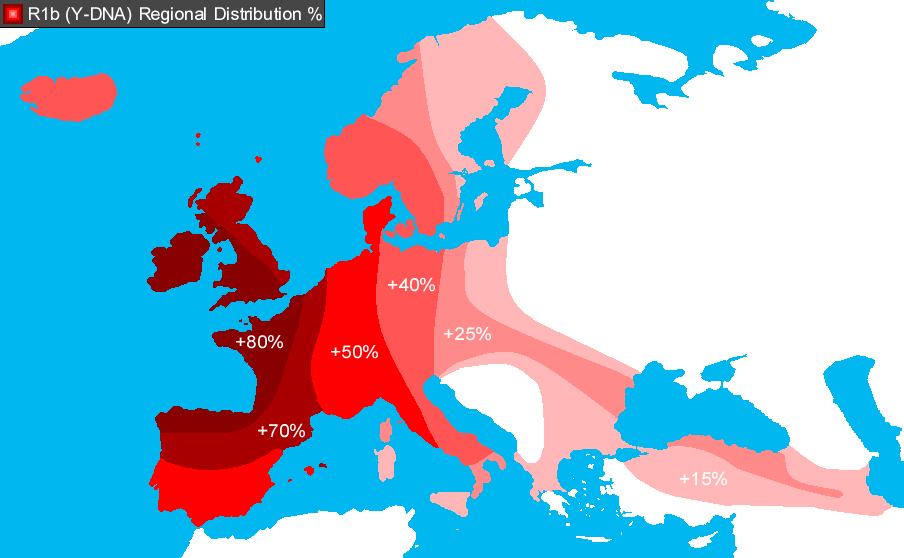
So much for the Afro-centrists and others who have derided the very obvious northwestern European appearance of a large number of the pharonic mummies. It seems like March of the Titans was right after all…
RELATED ARTICLES
- Researchers Warn Vaping is just as Harmful as Cigarettes
- DNA test to be added to Covid screening
- Justice in Corrupt Germany: 8 out of 9 Immigrants who Gang Raped Minor Girl in Park ON VIDEO, set Free
- FDA says Targeted Gene Therapy may CAUSE Cancer instead of Curing it
- Canada Confirms Undisclosed Presence of DNA Sequence in Pfizer Shot










the Skeptic
What don’t YOU understand – your data was erroneous, and, outdated. Even one or YOUR earlier sources indicated J* does NOT occur in Blacks – YOUR SOURCE. Most of my links showing this, are very recent. Find other sources backing YOUR claim.
You are either being deliberately obtuse or you are just ignorant. I wrote about the ancestral allele 12f2.1 which is found in Black Africans :
l]http://hmg.oxfordjournals.org/content/9/17/2563.full[/url]
“…Based on these alignment data, the molecular origin of the 12f2 STS polymorphism is assumed to be linked to the deletion of the L1PA4 element in the HERV15yq2 sequence block. The polymorphic 12f2 Y-DNA marker was first described as a restriction fragment length polymorphism (RFLP) for EcoRI and TaqI in human male DNA (12). In genomic DNA blots of human MALES and FEMALES, the probe 12f2 (Fig. 1a) cross hybridized to a polymorphic male-specific EcoRI fragment of 5.2 and 3.2 kb, whose presence was mutually exclusive.
“…It seems likely that the presence of the L1PA4 element in the HERV15yq2 sequence block represents the ancestral state in humans since POPULATIONS WITH ONLY THE ‘LONG’ 12f2 ALLELES (5.1 kb EcoRI/10 kb TaqI) were FOUND IN AFRICAN BLACKS, in ORIENTALS and in NATIVE AMERICANS and a FREQUENCY GRADIENT OF ITS DELETION was observed in EUROPEAN populations (34).
The deletion is found in EUROPEANS. I do not care if you want to call Arabs Europeans….the point is the nomenclature is clear – African Blacks, Orientals, Native Americans and Europeans.
You blew it. Lebanese men have tested positive for 12f2b but negative M172 and M267, so it is not specific to White-Semitic (?) but Europeans.
the Skeptic
Sorry buddy, I did NOT blow it – f12f2a (deletion), from YOUR source, Quote,”…Middle Eastern haplogroup J, characterised by the mutation 12f2a (deletion), is also found frequently in the Amhara and Tigray people. It exists at levels of about 35% among the Amhara, of which about 33% is of the type J-M267, almost all of which was acquired during Neolithic times or earlier, while 2% is of the derived J-M172 type representing admixture due to recent and historic migrations.
What you are FAILING to understand is J* = 12f2a (deletion), a Semitic White haplomarker – the groups you listed, are in Arab populated regions, and, are heavily mixed Black/Arab peoples. Your source is old, and erroneous – the links I show, provide evidence J* (all subclades) originate in the Middle East, and, spread to Europe(including India, an, Pakistan), parts of Oriental Asia, and, parts of Black Africa – AGAIN, J does NOT occur in Blacks,or, any other ethnic group, EXCEPT Semitic Whites and MIXED peoples, in this case, Arab/Black mixes. Please, get some grounding in Genetics so you comprehend what you copy and past. _ I also showed you a quote from one of YOUR sources BACKING what I just said – NO J IN BLACKS.Do your self a favor – go to the Library, and research Genetics. I am growing tired of proving you wrong, even using your OWN sources.
You obviously have a blind spot….
[url]http://hmg.oxfordjournals.org/content/9/17/2563.full[/url]
“…Based on these alignment data, the molecular origin of the 12f2 STS polymorphism is assumed to be linked to the deletion of the L1PA4 element in the HERV15yq2 sequence block. The polymorphic 12f2 Y-DNA marker was first described as a restriction fragment length polymorphism (RFLP) for EcoRI and TaqI in human male DNA (12). In genomic DNA blots of human MALES and FEMALES, the probe 12f2 (Fig. 1a) cross hybridized to a polymorphic male-specific EcoRI fragment of 5.2 and 3.2 kb, whose presence was mutually exclusive.
“…It seems likely that the presence of the L1PA4 element in the HERV15yq2 sequence block represents the ancestral state in humans since POPULATIONS WITH ONLY THE ‘LONG’ 12f2 ALLELES (5.1 kb EcoRI/10 kb TaqI) were FOUND IN AFRICAN BLACKS, in ORIENTALS and in NATIVE AMERICANS and a FREQUENCY GRADIENT OF ITS DELETION was observed in EUROPEAN populations (34).
I did not know there was a statute of limitations on infertility studies, since when?
None of the urls you posted speak to the 12f2.1 allele in any detailed way. You blew it. The allele is a factor of human male and female DNA period.
The Skeptic
All J* originates in the Middle East, spreading out from there, INCLUDING J-M267 – sorry, but YOU are in error, as is your information – my links are recent, and, include experts – the presence of J* is in Semitic peoples, and MIXED peoples – in this case, Arabs/Blacks – it simply does NOT occur in Blacks: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1181965/
http://www.familytreedna.com/public/J1_asterisk_Y-DNA/default.aspx?section=ysnp
http://dienekes.blogspot.com/2009/09/y-chromosomes-of-saudi-arabia.html
http://en.wikipedia.org/wiki/Haplogroup_J1e_%28Y-DNA%29
http://dienekes.blogspot.com/2009/09/y-chromosomes-of-saudi-arabia.html
http://www.freebase.com/view/en/haplogroup_j1
http://www.nature.com/jhg/journal/v56/n1/full/jhg2010131a.html
http://hpgl.stanford.edu/publications/AJHG_2004_v74_p1023-1034.pdf
http://www.servinghistory.com/topics/Haplogroup_J1c3_%28Y-DNA%29
CN,
You are incorrect the 12f2b (12f2.2)deletion is found only in Europeans.
You are incorrect in that the 12f2a (12f2.1)(Undeleted) is found in African Blacks, Orientals, i.e., Near Easterners, and Native Americans.
L1PA4 deletion:
The deletion of the L1PA4 element was recognized as the molecular origin of the DYS11 12f2 restriction fragment length polymorphism.
http://docs.google.com/viewer?a=v&q=cache:nBYGCDu-6mUJ:www.pasteur.edu.uy/publico/bonilla/Genetica%2520Humana/Fathers%2520and%2520sons%2520The%2520Ychromosome%2520and%2520Human%2520Evolution.pdf+Ann+Hum+Genet+54.287-296&hl=en&gl=us&pid=bl&srcid=ADGEESi9A5pSglrYzCGUBaMt5xxXIbrf7ckuq0Ek3ErT-pB0yGclDYhjSJ3bIz1npB50n15rtXgrNPQoBnpBugjxYIqszXIoKE_lHWmACCymrjuAwbKcim0FYfJCYm_nyy-illqexQI2&sig=AHIEtbTpdaSJjaDNuOHhRh9LcEoS4U4s0w
“The geographical distribution of loci, such as YAP, focuses attention on specific novel questions about population origins. Although the Alu insertion is found at highest frequency in Africa, it is found at moderate frequency in Japan and at low frequency in some other areas, such as Europe. Its presence outside Africa is often not due to recent admixture, but does it represent, for example, admixture a few centuries ago or the presence of YAP* chromosomes in founding populations? These questions cannot be answered yet, but more detailed analysis of the Y chromosomes should allow such possibilities to be distinguished.”
Recap :
(1) The 12f2-b (12f2.2)DELETION is found only in Europeans.
(2) The 12f2-a (12F2.1)UNDELETED is found in African Blacks, Orientals, i.e., Near Easterners, and Native Americans.
:oops: Deletions in Y-DNA Haplogroup R1…(Europeans)…
Page 99:
http://books.google.com/books?id=IIi2HSkHn1AC&pg=PA98&lpg=PA98&dq=J*12f2+deleted?&source=bl&ots=GFqKg8SbkK&sig=MaAE4TEjN-JnTttVXRiLYRm9evw&hl=en&ei=8qx0Tp_6FoyTtweGk926DA&sa=X&oi=book_result&ct=result&resnum=4&ved=0CDMQ6AEwAw#v=onepage&q=J*12f2%20deleted%3F&f=false
and finally…
AZF DELETIONS AND Y CHROMOSOMAL HAPLOGROUPS : HISTORY AND UPDATE BASED ON SEQUENCE, PETER H. VOGHT, Section of Molecular Genetics & Infertility, Department of Gynecological Endocrinology & Reproductive Medicine, University of Heidelberg, Heidelberg, Germany
http://humupd.oxfordjournals.org/content/11/4/319.full.pdf
“…However, the two variable fragment lengths (8, 10.4kb) observed after TaqI restriction were only present in genomic male DNA, indicating their Y chromosomal origin (Figure 2).
…Analysis of the frequency of this 2.4kb deletion polymorphism (here designated as DYS11 12f2-2.4kb allele) in different populations from Europe, Africa and Asia revealed it SPECIFICITY for Caucasian populations because it is ABSENT in African BLACKS, in ORIENTALS and in NATIVE AMERICANS (Semino et al., 1996).
…Most interesting, the frequency of the 12f2-2.4kb deletion decreased from the Near East to northwestern Europe populations, reflecting the neolithic demic diffusion of the ancient farming cultures.
…Today we know that the 12f2-2.4kb deletion is the derived state (12f2.2/12f2-B allele) of the undeleted ancient DYS11 sequence (12f2.1/12f2-A allele) and that this deletion must have occurred at least two times during the evolution of the human population history (Blanco et al., 2000).”
Resolved :
The 12f2-a (12f2.1)UNDELETED is found in African Blacks, Orientals, i.e., Near Easterners, and Native Americans.
Repeated Y-chromosome degeneration in human evolutionary genomic events!
J*12f2.1 /12f2a – Not admixture!
Y-DNA Hg BT Mutations : SRY108301.1G+, SRY10831.2A+. Ancestral 12f2.1 (12f2a-10kb.- undeleted.
Y-DNA Hg D2 Mutations : M57, M64.1, M179, P37.1, P41.1, P190, M55. Deletion: 12f2b, 12f2.2-2.4kb (deletion)
Y-DNA Hg J Mutations : M304, P209, L60, L134. Insertion = present: 12f2a, 12f2.1-10kb.
In fact studies have shown that men can be positive for 12f2.1 and negative M172 and M267 !
http://www.members.cox.net/generalbanks/completecategorieshapj.html
CN,
you wrote:
“…it does NOT occur in Blacks(Sub Saharan), just Semitic White populations- END TRANSLATION.
…Middle Eastern haplogroup J, characterised by the mutation 12f2a (deletion), is also found frequently in the Amhara and Tigray people. It exists at levels of about 35% among the Amhara, of which about 33% is of the type J-M267, almost all of which was acquired during Neolithic times or earlier, while 2% is of the derived J-M172 type representing admixture due to recent and historic migrations.”
Well, it has taken some time to get the RIGHT answer, but this is the genetic gist of allele 12f2.1/12f2.2:
[url]http://hmg.oxfordjournals.org/content/9/17/2563.full[/url]
“…Based on these alignment data, the molecular origin of the 12f2 STS polymorphism is assumed to be linked to the deletion of the L1PA4 element in the HERV15yq2 sequence block. The polymorphic 12f2 Y-DNA marker was first described as a restriction fragment length polymorphism (RFLP) for EcoRI and TaqI in human male DNA (12). In genomic DNA blots of human MALES and FEMALES, the probe 12f2 (Fig. 1a) cross hybridized to a polymorphic male-specific EcoRI fragment of 5.2 and 3.2 kb, whose presence was mutually exclusive.
“…It seems likely that the presence of the L1PA4 element in the HERV15yq2 sequence block represents the ancestral state in humans since POPULATIONS WITH ONLY THE ‘LONG’ 12f2 ALLELES (5.1 kb EcoRI/10 kb TaqI) were FOUND IN AFRICAN BLACKS, in ORIENTALS and in NATIVE AMERICANS and a FREQUENCY GRADIENT OF ITS DELETION was observed in EUROPEAN populations (34).
We assume that a gene conversion event between the two HERV15yq sequence blocks in proximal Yq11 most likely caused the precise deletion of this L1PA4 element.”
[url]http://docs.google.com/viewer?a=v&q=cache:H4w_tkJUJS0J:www.ncbi.nlm.nih.gov/pmc/articles/PMC1757162/pdf/v037p00752.pdf+12f2+deletion+%2B+Ethiopia&hl=en&gl=us&pid=bl&srcid=ADGEESiJBKwzi5wG4Iz_fB78DbiVS5fylAYAk6ufu2RHJ68VxxeDb_YelwWSCLsV1NV1bC9XkT-PC6OgoibMllfKHmEq04fvRIY7FvJ7M71mAXNUJsRe3RWcaOrvzR1yWEy0RB0YnbwI&sig=AHIEtbRwiuZ60SAo0ZwmwDtTduH3rEi0FA[/url]
“…The assumption that markers such as the 12f2 deletion are neutral is required for their use in reconstructing human prehistory. This assumption may be violated if these markers represent permutations, though only if negative selection outweighs drift.”
the Skeptic
Actually – the answer is from the same article YOU just gave, QUOTE,”The J1 subclade is found in Africa, but limited to the North and Northeast corner (Horn of Africa) adjacent to Sinai and Arabian Peninsulas. *****It is not found in Sub-Saharan populations. Semitic- and Caucasian-language populations in Africa have a typically high frequency of J1.***** – EMPHASIS mine – TRANSLATION – it does NOT occur in Blacks(Sub Saharan), just Semitic White populations- END TRANSLATION. Examination of the results for the phylogeography of subclade J1 and its STR variation support two African migrations – an earlier one during the Neolithic era driving Southward toward Ethiopia and a later one (7th century AD) traveling North. The second migration may have been spread by Arab slave trade, perhaps Bedouin groups. Further propagation was likely provided by Muslims in 6th century AD.”
Middle Eastern haplogroup J, characterised by the mutation 12f2a (deletion), is also found frequently in the Amhara and Tigray people. It exists at levels of about 35% among the Amhara, of which about 33% is of the type J-M267, almost all of which was acquired during Neolithic times or earlier, while 2% is of the derived J-M172 type representing admixture due to recent and historic migrations.
Unlike their Y-DNA, Ethiopians’ mtDNA is more typical of sub-Saharan Africans, leading researchers to conclude that “Caucasoid gene flow into the Ethiopian gene pool occurred predominantly through males. Conversely, the Niger-Congo contribution to the Ethiopian population occurred mainly through females.”:http://en.wikipedia.org/wiki/People_of_Ethiopia
Bottom line – Ethiopians have much Black/Semitic White ADMIXTURE – just what point are you attempting to make – NOTHING states all peoples are descended from Black people – if that is your point – I have spent an inordinate amount of time, proving you INCORRECT – even using YOUR own copy/paste. Arabs, Jews, and Blacks(closest to North Africa – part of the ME) have much ADMIXTURE.
The Y-DNA Haplogroup J story …so far : http://www.dnaancestry.ae/Y-DNA-Haplogroup-J.php
Early origins
The origin of Y-DNA Haplogroup J maps to the Middle East around the ‘Fertile Crescent’, an area also known as the ‘Cradle of Civilization’ since this area saw the birth of many technological advancements that helped humans move from nomadic hunter-gatherers to an agriculture-based society living in one place. The sprouting of some the first cities and empires in human history were contingent on these developments and featured the proliferation of Haplogroup J.
The precise location for the origin of Haplogroup J is not known, but its prominence in the Near East/West Asia and the Middle East/Central Asia indicates that it likely arose in one of these regions. It is closely associated with the Fertile Crescent; an area spanning the Nile and Tigris/Euphrates River systems, with the Levant (present day Lebanon) in between. This region has encompassed many early cultures and empires from the Stone Age (Neolithic) to the Iron Age and has also been dubbed the ‘Cradle of Civilization’. Societies, dynasties and empires in this broad region include the Sumerian, Assyrian, Babylonian, Egyptian, Phoenician and Persian. Haplogroup J is also particularly abundant in Anatolia (present day Turkey) and the Y-chromosome diversity observed here suggests that this area is a possible source of this clade. Owing to these strategic locations, Y-DNA Haplogroup J is common on three continents: Asia, Europe and Africa.
Y-DNA Haplogroup J is a descendent of suprahaplogroup F, which encompasses a large group Y-DNA lineages (haplogroups F-T, see Figure 3). Suprahaplogroup F is believed to have migrated from Africa approximately 50kya. Haplogroup J arose approximately 30kya (see Figure 4) and has been defined by a number of unique Y-chromosome polymorphisms; the 12f2a deletion and the M304 and P209 SNPs.
Before I go any further, tell exactly what you think the 12f2a deletion tells us about the divergence of J* into J12f2a. Exactly. Not your opinion. What does this deletion tell us?
If you do not know, say so.
the Skeptic
Actually the J1 haplogroup IS found in HIGHER amounts in Egypt than what YOU listed: http://en.wikipedia.org/wiki/Haplogroup_J_%28Y-DNA%29 and http://www.dnaancestry.ae/Y-DNA-Haplogroup-J.php
I think we agree, the J haplogroup originates in the ME – Arab and Ethiopian clashes explain the High J1 and J2 in Ethiopia as well as the Northern 1/3 of Ethiopia being Arab, NOT, just by language – ethnicity – the DNA evidence both you, and, I presented – shows Semitic White (most likely, BOTH, Arabs, and Jews) influence in admixture with the Ethiopians. Seems you are attempting to say, in a round about way, Ancient Egyptians were the Black Nubians – they were NOT – they were Arab Semitic White – the J haplogroup only occurs in Arabs, Jews, and MIXED peoples – bottom line – many Ethiopians, Somalis, Sudanese, Egyptians, are a mixed people.
CN
Y-DNA J12f2a – J12f2.1 : Genetic Equivalents L134, M304/Page16, P209, S6/L60, S34, S35 :
Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism.
http://jmg.bmj.com/content/37/10/752.full
Polymorphic : The occurrence of different forms, stages, or types in individual organisms or in organisms of the same species, independent of sexual variations.
“Another marker is the 12f2 marker, which is located on the long arm of the Y-Chromosome on the AZFa region and was shown to be polymorphic among populations.
Recently, B lanco et al., developed a new PCR assay and screened a group of diverse Y-chromosomes for the 12f2 polymorphism. Their results revealed that the 12f2 amplicon was absent from Y-chromosomes, whose haplotype backgrounds are defined by the presence of the YAP insertion, and they suggested that such deletion polymorphisms may have occurred more than once during human evolutional history (22). The 12f2 deletion is found at the highest frequencies (greater than 25% in Middle Eastern, Southern European, North African and Ethiopian populations (23).
“Although it was reported that the YAP+ chromosomes from Africa and Japan are identical by descent (7,15), the present results of the D FFRY and 12f2s markers shows that the Japanese YAP+ chromosomes are different from YAP+ chromosomes of Bolivian, African-American, and Caucasian-American males. Hence, we support the theory that “different populations often have characteristically different Y-chromosomes.”
Y-DNA J12f2a – J12f2.1 : Genetic Equivalents L134, M304/Page16, P209, S6/L60, S34, S35 :
“Today the M304 marker appears at its highest frequencies in the Middle East, North Africa and Ethiopia. In Europe M304 is seen only in the Mediterranean region.”
Y-DNA J1c3 : “The P58 marker which defines subgroup J1c3 is common, and was first announced in Karafet et al. (2008), but had been announced earlier under the name “Page08” in 2006 in Repping, Van Daalen & Brown (2006).[4] This haplogroup dominates haplogroup J, and is notable as containing the Jewish “Cohen modal haplotype”, as well as both the so-called “Galilee modal haplotype” and “P&I Arab modal haplotype” associated with Palestinians and Israeli Arabs by Nebel et al. (2000).[7]
More generally, the J-P58 group has been shown to be closely associated with a large cluster of J1 which had been recognized before the discovery of P58. This cluster was identified by STR markers haplotypes – specifically YCAII as 22-22, and DYS388 having unusual repeat values of 15 or higher, instead of 13.[4] Apart from the Jewish “Cohen” haplotype, Semino et al. (2000) associated this DNA profile with the Arab expansion in the seventh century AD, and noted that it was most frequent amongst J1 men in the Middle East and North Africa, but less frequent in Ethiopia and Europe.
Similarly, Tofanelli et al. (2009) refer to an “Arabic” and a “Eurasian” type of J1. The Arabic type includes Arabic speakers from Maghreb, Sudan, Iraq and Qatar, and it is a relatively homogeneous group. This is the group with YCAII=22-22 and high DYS388 values. The more diverse “Eurasian” group includes Europeans, Kurds, Iranians and Ethiopians (Ethiopia being outside of Eurasia), and is much more diverse. The authors also say that “Omanis show a mix of Eurasian pool-like and typical Arabic haplotypes as expected, considering the role of corridor played at different times by the Gulf of Oman in the dispersal of Asian and East African genes.”
That takes care of the other possible genetic link to the ‘Jews’ besides E-M35.
Not Ancestry but Chance Mutations:
“While unusual Y-STR values can lead to clues of common ancestry, caution should be used when utilizing a single Y-STR marker or a small group of Y-STR markers to infer ancestry as false associations can arise. This situation is illustrated by Y-STR marker DYS458, which displays an unusual “0.2” value (ex. DYS458 = 17.2) in all haplogroup J1-M267 haplotypes observed to date. Consequently, “0.2” values have been used to infer membership within haplogroup J1-M267. However, DYS458.2 values have recently been found to also exist within haplogroups R1b1b2-M269 and R1b1b2a1-M405. Haplogroups J and R do not share this unusual marker because of common ancestry but due to chance independent mutation events. This result underscores the weakness of using a single STR marker or too few STR markers as indicators of common ancestry, even when those values are relatively rare.”
http://www.genetree.com/y_j
Y-DNA J1/J2:
The question is not why the Ethiopians have Y-DNA J1 but, why Egypt does not have more?
In other words, if Y-DNA J1 or J2 and its various clades or subclades represent the Arab as the ancient Egyptian, why there is not more in Egypt, but more in Sudan and Khartoum and Yemen, South Arabia.
I have given you a link to the distribution of J1c3
http://s4.zetaboards.com/Afro_Asian_mt_DNA/topic/9039225/1/#new
Ethiopian Omoro – sample size of 78 men – 2.6%.
Ethiopian Amhara – sample size of 48 men – 29.20%.
Egypt – sample size 147 males – 21.10%.
Khartoum, Sudan – sample size 35 males – 74.30%
Yemen – sample size 62 males – 72.6%.
That is the question. Where is the ancient presence of J1 most dominant? Khartoum -not Egypt.
Why?
CN
Y-DNA E(M35):
Origins of Falasha Jews studied by Haplotypes of the Y chromosome : http://findarticles.com/p/articles/mi_qa3659/is_199912/ai_n8872400/
“Abstract DNA samples from Falasha Jews and Ethiopians were studied with the Y-chromosome-specific DNA probe p49a to screen for Taql restriction polymorphisms and haplotypes. Two haplotypes (V and XI) are the most widespread in Falashas and Ethiopians, representing about 70% of the total number of haplotypes in Ethiopia.
…Because the Jewish haplotypes VII and VIII are not represented in the Falasha population, we conclude that the Falasha people descended from ancient inhabitants of Ethiopia who converted to Judaism.”
Now, if you can read…..
“…the Jewish haplotypes VII and VIII are not represented in the Falasha population, we conclude that the Falasha people descended from ancient inhabitants of Ethiopia who converted to Judaism.”
African Converts to Judaism:
“Unsurprisingly, quite a number of genetic studies have been conducted to clarify the status of Falasha people. Already in the early 1990s, a study of Y-DNA and mtDNA of Ethiopian Jews has been conducted and reported in Zoossmann-Disken et al. (1991) who found that
“Ethiopian Jews cluster with other Ethiopian tribes and occupy a central position on a principal component map between African and Asian populations.”
LUCOTTE & SMETS:
“A further study by Lucotte & Smets (1999) came to a similar conclusion: although they do not doubt the Jewish practices of Falasha, their claim to a descent from the tribe of Dan is shown to be wrong:
“… the distinctiveness of the Y-chromosome haplotype distribution of Beta Israel Jews from conventional Jewish populations and their relatively greater similarity in haplotype profile to non-Jewish Ethiopians are consistent with the view that the Beta Israel people descended from ancient inhabitants of Ethiopia who converted to Judaism.”
HAMMER et al.:
“Similarly, Hammer et al. (2000) treat the Beta Israel as an EXCEPTION to the general commonality of the “paternal gene pools of Jewish communities from Europe, North Africa, and the Middle East descended from a common Middle Eastern ancestral population…” — the Falasha are “affiliated more closely with non-Jewish Ethiopians and other North Africans.”
Other Genetic links?
“But a year later, an interesting twist appeared in the story of the Falasha: a study conducted in 2001 by the Department of Biological Sciences at Stanford University found a possible genetic similarity between 11 ETHIOPIAN Jews and 4 YEMENITE Jews. So are Ethiopian Jews related to other Jewish groups worldwide by blood after all? Probably not: the more likely explanation proposed by the Stanford researchers is that there existed a gene flow between Ethiopian and Yemenite Jewish populations, or perhaps even between Jewish and non-Jewish populations of both regions. Thus, it is not that the 11 Ethiopian Jews studied are related to other Jews by blood, but rather the four Yemenite Jews are related to Ethiopians (perhaps they are descendents of reverse migrants of African origin who crossed the Red Sea from Ethiopia to Yemen).”
Y-DNA studies and mt-DNA studies to determine ‘Jewish’ biologogical ancestry:
“Note that these earlier studies typically focused on Y-DNA, which traces paternal descent. But what about women and maternal descent? After all, for many strands of Judaism today it is the MOTHER’s LINEAGE that is important in determining whether a person is Jewish or not.
A more recent study, reported in Thomas et al. (2002) looked specifically at the mtDNA (mitochondrial DNA) of Ethiopian Jews. According to their results, the most common mtDNA type found among the Ethiopian Jewish sample was present only in Somalia, which further supports the view that most Ethiopian Jews are of local, Ethiopian origin.”
The Falasha are not mixed, admixed but of the same Y-DNA and mtDNA of the local Ethiopian population and this substantiates the degree of admixture.
I am done with this. I will give you just one more post for Y-DNA J1-172, etc.
the Skeptic
I NEVER siad the Black Ethiopan Jews are White, or, Arabized – I SAID, MIXED, and,presented evidence of such through patrilinear studies – Semitic White Jewish males(probably Arab Semitic White males) took Black Ethiopian wives – I presented evidence the J haplogroup ORIGINATES in the ME(Near East) – now if you can NOT accept admixture – YOU have a problem. Some of the Falasha are MIXED – some are by religious conversion. From the interaction of the two, or, three ethnic groups, that is a very logical concluusion, BACKED by studies. If the J haplogroup originates in the ME – two ways it got in Ethiopian Blacks – they went to the ME – Ethiopian Enpire in 1500 B.C. or, Arabs, and/or Jews went to them- Arabs dove the Ethiopians BACK to Ethiopia, and, King Solomonn, and, his men visited Ethiopia. For some reason, you can NOT accept this – even your copying/ pasting explains it this way – ADMIXTURE – just what is your propblem – besides – the Northern 1/3 of Ethiopis is CURRENTLY Arab – has been since ancient times.
I have not written about Haplogroup D…so…
I was writing about Y-DNA-E-P2/Pn2 (E-M35).
I am also tired of this subject and how you insist on making Ethiopians either White-Semitic-Jews or ‘White-Semitic-Arabs.’
If you cannot understand that M35 split off from the Eastern Africa/Eastern Africa. – Afrasians, Afro-Asiatic, in the Near East and entered with different SNPs, clades, etc, then I am not the one who has the problem..
‘Africans who are P2(E-M35), PN2(E-M35), and the clades that are also ‘African’ specific SNPs will be genetically joined regardless of ‘color’.
“The modern population of E-M215 and E-M35 lineages are almost identical, and therefore by definition age estimates based on these two populations are also identical. E1b1b (E-M215) and its dominant sub-clade E1b1b1 (E-M35) are believed to have first appeared in East Africa about 22,400 years ago.”
I am done.
the Skeptic
Actually, for the most part, you got Abraham,Hebrew, Arab, and Jew correct – Jew is BOTH an ethnic classification as in Semitic White, and, by religious conversion. This applies to the Falasha Jews – most wer by religious conversion – thus, truly all Black, BUT King Solomon, and, his men,had relations with Black Ethiopian women, either by force, or, consensual – those Unions produced mixed children – part Black,/Part Jewish Semitic White. because of no standardization of the constantly changing genetic terms – there is much confusion. As to the origin of haplotype V – whether it originated in Africa, or, the Middle East – is divided: http://en.wikipedia.org/wiki/Haplogroup_E1b1b1b_%28Y-DNA%29 and http://en.wikipedia.org/wiki/Haplogroup_E1b1b1b_%28Y-DNA%29
Haplo group D, a precursor to M35, does NOT occur in Africa, so, although higher percentages are found in parts of Africa( also in North Africa, among the Arabs)., it is NOT certain that Africa is the origin, but a back flow from the Middle East. Please, state clearly what your point is – my impression is either you are claiming Semitic peoples are Black, or, that Black peoples are the credit for the achievements Semitic White(Arabs, and, Jews)peoples claim. For the record, it is NOT known what ethnicity the Queen of Sheba was – Black, or, Arab Semitic White – she ruled BOTH at the same time.
CN
You stated…
“I can NOT explain it more clearly than I already have – i posted the patrilinear study showing Jewish lineage in some of TWICE, I explained theAbraham – Arab- Jew split TWICE, and you still either don’t accept it, or, are not comprehending it. I really have nothing to add to this, other than to tell you go back, and, read my answers slowly, then, THINK on them. On the Falasha – some have Jewish heritage – apparently, most are by religious con version.They, being genetically, slightly different genetically than other Ethiopians of course, can be explained as admixture, and/or genetic mutation – since it is limited, my personal opinion is admixture, probably BOTH, Arab, and, Jewish – since the Northern 1/3 of Ethiopia, is Arab, AND, Muslim(don’t eat pork) – Arabs is the most likely source.”
“…my personal opinion is admixture, probably BOTH, Arab, and, Jewish – since the Northern 1/3 of Ethiopia, is Arab…
THAT is A BLATANT LIE: Chiaroni et al. (2010)
Ethiopia Amhara 48 Males tested.
29.2% – J1.
8.3% – J1 but not P58.
20.8% – J-P58 (J1c3).
Ethiopia Y-DNA J1 :
“…Chiaroni et al. (2010) agree with Tofanelli et al. (2009) in doubting that the Islamic expansions are old enough to completely explain the patterns of J1 frequencies, especially in areas such as Turkey, the Caucausus, and Ethiopia. These areas are notably high in J1* (see below under distribution) and are also regions where the Neolithic arrived with people from the Near East, but they WERE NOT ARABIC SPEAKING.”
:oops: The ‘Berber Marker’ subhaplotype Vb [in this study] :
http://findarticles.com/p/articles/mi_qa3659/is_200606/ai_n17175647/pg_2/?tag=mantle_skin;content
“Subhaplotype Vb is the Berber subhaplotype because its most elevated relative value (63.5%) is obtained for the Berber population of Marrakech.
In the non-Berber population of Rabat in Morocco, the frequency of subhaplotype Vb is only 20.6%, whereas the frequency of subhaplotype Va (Arab) is 37.3%.
In order of decreasing values, the subhaplotype Vb frequencies are
40% in Mauritania or 27 males tested,
35.9% in South Portugal or 24 males tested,
25.4% in Andalusia or 17 men tested, and
15.8% in Libya or 10 males tested.
Low frequencies of subhaplotype Vb are found in Sicily (5.1%), Algeria (2.8%), Tunisia (2.7%), and North Portugal (2.5%); frequencies less than 2% are found in French Basques (1.9%), in Naples (0.8%), and in Corsica (0.6%), Subhaplotype Vb is absent in Catalonia (Barcelona and Perpignan), in the south of France (Montpellier, Grasse, and the region of Marseilles), in continental Italy (Milan, Genoa, and Rome), and in Sardinia.”
“In our PCR assay the 68 Moroccan subjects with subhaplotype Vb
(47 Berbers and 21 Arabs)
were tested for the M81 marker: All subjects were positive for the M81 marker, so subhaplotype Vb is homologous with subhaplogroup E3b2.
The 38 Moroccan non-Berber subjects were further tested for the M78 marker: Only 31 of them (80.8%) were positive for the M78 marker; we conclude that, in Morocco at least, subhaplotype Va corresponds only partly to subhaplogroup E3b1.”
E1b1b1b1 (E-M81). Berbers, but also Spain, France, Italy, Turkey, etc.
AGAIN…You keep adding to or changing your position and are still pushing this…
“…On the Falasha – some have Jewish heritage – apparently, most are by religious con version. They, being genetically, slightly different genetically than other Ethiopians of course, can be explained as admixture, and/or genetic mutation…”
Now, suddenly you see the light?
– Some have Jewish heritage
– Some were converts
– Some slightly different genetically than other Ethiopians.
– Some can be explained as admixture or genetic mutation.
Well, admixture is not the same as genetic mutation.
So, to be clear on what I have been saying,
Most Falasha, ‘Black Jews’ are no different genetically than the local Ethiopian population : Y-DNA E-P2/PN2(M35), mt-DNA L.
Dag.
That study was of 48 Ethiopians and only 14 Amhara samples were J1 or 29%. Do you honestly think any genetic study has been done for the ENTIRE POPULATION OF NORTHERN ETHIOPIA?
Religions and Demographics of Amhara:
“Based on the 2007 Census conducted by the Central Statistical Agency of Ethiopia (CSA), the Amhara Region has a population of 17,214,056 of whom 8,636,875 were men and 8,577,181 women; urban inhabitants number 2,112,220 or 12.27% of the population. With an estimated area of 159,173.66 square kilometers, this region has an estimated density of 108.15 people per square kilometer. For the entire Region 3,953,115 households were counted, which results in an average for the Region of 4.3 persons to a household, with urban households having on average 3.3 and rural households 4.5 people. At 91.48% of the local population, the region is predominantly inhabited by people from the Semitic-speaking Amhara ethnic group. Most other residents hail from other Afro-Asiatic communities, including the Agaw/Awi (3.46%), Oromo (2.62%), Agaw/Kamyr (1.39%) and Argobba (0.41%).
Of the total population of the region, 82.5% were Orthodox Christians, 17.2% Muslim, 0.2% Protestants, and 0.1% practiced other faiths.”
http://en.wikipedia.org/wiki/Amhara_Region
8,636,875 Men!!! So even if we use the 29% J1* for the Amhara that is about 2,504,693 Men – hardly an ‘Arab’ genetic majority, if the SNPs are even ‘young’ enough to correlate to the Arab influx.
Compared to the 80% of the population who are Christians (14.1 million) to those who are Muslim (2.9 Million) well…you do the math.
The effect of Amharization:
“At times over the centuries, the Amhara have been attacked by military forces led by Muslim leaders, carving out their own political territory and imposing Islam on the conquered.
…The Amhara have an important Christian heritage and have kept their faith in the face of great opposition at some periods in their history.
Only a few Amhara are Muslim. Traditionally Muslims have been considered invaders and enemies, so Islam has not had much appeal to the Amhara.”
So, in future, leave your opinion out and stick to the facts. The Amhara are ‘Black-Semitic-Africans’ who speak Amharic – their Semitic language and practice Christianity and not Islam. They are not ‘Arabized’.
Falasha Jews? Just read the previous paragraph and substitute where appropriate.
http://s4.zetaboards.com/Afro_Asian_mt_DNA/topic/9025811/1/
I hope this ends the debate, finally:
mtDNA of Yemeni and Ethiopian Jews
From the paper: http://dienekes.blogspot.com/2010/07/mtdna-of-yemeni-and-ethiopian-jews.html
“Mitochondrial DNA analysis also revealed a high diversity of sub-Saharan African and Eurasian haplotypes in both the Yemenite and Ethiopian Jewish populations (see Fig. 2). Specifically, common haplotypes (haplotypes present at [5%) in Yemenite Jews include the African haplogroup L3x1 and Eurasian haplogroups R0a (renamed from (preHV)1 (Torroni et al., 2006), HV1, J2a1a [renamed from J1b (Palanichamy et al., 2004)] K, R2, U, and U1, and in Ethiopian Jews include African haplogroups L2a1b2 and L5a1 and Eurasian haplogroups R0a and M1a1 (see Fig. 2). Overall, sub-Saharan African L haplotypes [hereafter referred to as L(xM,N), i.e., all African haplotypes except M and N, following the nomenclature of Behar et al. (2008)], comprise a large proportion of the genetic variation in both Jewish populations, representing 20% in the Yemenite Jews and 50% in Ethiopian Jews. This high frequency contrasts with other Jewish populations, such as Near Eastern and Ashkenazi Jews, who almost entirely lack L(xM,N) haplogroups (Thomas et al., 2002; Richards et al., 2003).”
They are descending from a non Semitic mtda they are not “true Hebrews” for the orthodox Judaism but converts.
Next…
CN
Really. I am just trying to understand why Abraham is A Hebrew but not a ‘Jew’ but all ‘Jews’ are descended from him. So what is the difference between a Hebrew and a ‘Jew’.
If you cannot explain it reasonably then the conclusion is you don’t know what you claim you know and are just talking to talk the same old talk that people talk when they don’t know something they say they know. You know?
If, one means that ‘Jews’ are followers of Judaism, then anyone – anywhere – and of any ‘race’ can be a ‘Jew’. However, not all ‘Jews’ are Hebrew, and not all ‘Jews’ are Israelites, i.e. genetically descended from Abraham, Isaac, Judah, etc.
If, on the other hand, A Hebrew is a ‘Jew’ then are all Jews Hebrews regardless of their actual genetic-less link to Abraham?
If that is true, then not all Jews are Hebrews having not been born ‘Hebrew’ and as converts to Judaism are Hebrews in spirit or by spiritual proxy and the fulfillment of the covenant of circumcision.
Yet, the ‘Black Jews’, the Falasha are not direct genetic descendants of Abraham and as converts are Jews following Judaism. No one who is follower is automatically a Hebrew. Abraham was told his ‘seed’ would continue through Isaac the Hebrew and not Ishmael, the son of the bondwoman, Hagar the “Egyptian’.
That is the split.
You have Hebrew and Egyptian.
Judaism is a religion just like Christianity, Islam, Buddhism, etc, and and Hebrew or Egyptian can be a follower of any one of these religions.
Note:
“Throughout its history the community have been called a large number of names. “Beta Israel” originated in the 4th century, when the community was so named because they converted to Christianity during the rule of Ezana of Axum, the first monarch of the Aksumite Empire to embrace Christianity. This name wasn’t originally attributed any negative meanings and the community have used it ever since as their official name. Since the 1980s it has also become the official name used in the scientific literature to describe the community.
After the rise of the Solomonic dynasty (1270 AD), the Beta Israel community was also called Ayhudi (“Jews”) until they were referred to by the derogatory term Falasha (Ge’ez: “foreigners/exiles”), a term given to them by the emperor Yeshaq I of Ethiopia.”
:oops: Demographics and Semitic Languages : http://en.wikipedia.org/wiki/Axum
“Based on figures from the Central Statistical Agency in 2005, Axum has an estimated total population of 47,320 of whom 20,774 are men and 21,898 women.[11] The 1994 national census reported a total population for this city of 27,148, of whom 12,536 were men and 14,612 were women. The largest ethnic group reported was the Tigrayan (98.54%) and Tigrinya was spoken as a first language by 98.68%. The majority of the population practiced Ethiopian Orthodox Christianity with 85.08% reported as embracing that religion, while 14.81% were Muslim.[12].
Ethiopia and The Kingdom of Kush :
“Whatever the original character of this civilization, by the third century B.C. we are clearly dealing less with external influences and more with indigenous developments, which grow out of this earlier foundation. The language of local inscriptions, for example, is less and less like the original South Arabian language. Political terminology also appears less directly connected to South Arabian models. At the same time, cultural and trading ties seem to have developed with the Nile Valley, particularly the Nubian Kingdom of Meroe.”
Ethiopia and the Bible
“The Land of Cush. We begin our search for the source(s) of the Jewish elements in Aksumite culture with a consideration of the land known to biblical writers as “Cush.” The Hebrew Bible contains some fifty references to Cush or Cushites, most of which are translated in the Septuagint as “Ethiopia.” Since the ancient Greeks used the term “Ethiopia” to designate any southern land inhabited by people with “burnt-faces,” this appears to be a generally satisfactory rendering of the Hebrew term.
:oops: Unfortunately, some scholars ignore this usage and seriously confuse mattes by simplistically identifying ancient Cush/Ethiopia with present-day Ethiopia. This misreading not only distorts the intention of the biblical (and later Greek) authors, but also, when applied to the question of Beta Israel origins, produces connections and loci of Jewish settlement where none existed….”
“In the Hebrew Bible, the term Cush refers to a number of different locations. In some cases, particularly the genealogical table in Genesis 10:6-10, it is extremely difficult to understand the passages‘ intent. In others, most notably Esther 1:1 (“From India to Cush”),it seems to mean simply the ends of the earth. :-o In most instances, however, the term “Cush” is used to designate the “Nubian kingdom which was situated along the Nile, south of Egypt.” Such references are especially common in the eighth and seventh centuries B.C., when the twenty-fifth or “Ethiopian” (i.e., Nubian/Sudan Na-Pa-Ta) dynasty ruled Egypt and its kings played a major role in the international politics of the period.”
Read more: The Beta Israel (Falasha) in Ethiopia: from earliest times to the twentieth …
By Steven Kaplan ; http://books.google.com/books?id=UBXX7ZiPcOIC&pg=PT19&lpg=PT19&dq=Falasha+%2B+Jews+%2B+controversy&source=bl&ots=5Jxsy5JDSi&sig=NXqa4jv0KVzT4npF_7j_aUAZ0-I&hl=en&ei=ZzNaTojTN9P-sQKKzoGZDA&sa=X&oi=book_result&ct=result&resnum=7&ved=0CEUQ6AEwBg#v=onepage&q=Falasha%20%2B%20Jews%20%2B%20controversy&f=false
If you are unable to put the pieces of the puzzle of the ‘Black Jews’ back together, it is because you do not want to see what history has already proven.
Y-DNA E-M35 : I refuse to rehash the Y-chromosome again and only reiterate its Eastern African origons.
http://s4.zetaboards.com/Afro_Asian_mt_DNA/topic/9016010/1/
the Skeptic
I can NOT explain it more clearly than I already have – i posted the patrilinear study showing Jewish lineage in some of TWICE, I explained theAbraham – Arab- Jew split TWICE, and you still either don’t accept it, or, are not comprehending it. I really have nothing to add to this, other than to tell you go back, and, read my answers slowly, then, THINK on them. On the Falasha – some have Jewish heritage – apparently, most are by religious con version.They, being genetically, slightly different genetically than other Ethiopians of course, can be explained as admixture, and/or genetic mutation – since it is limited, my personal opinion is admixture, probably BOTH, Arab, and, Jewish – since the Northern 1/3 of Ethiopia, is Arab, AND, Muslim(don’t eat pork) – Arabs is the most likely source.
CN
No incursion. No admixture. Both the Falasha ‘Jews’ and the general local Ethiopian population SHARE Y-DNA E-M35 and mt-DNA L.
Dag.
This substantiates the extent of the admixture!!!! There is none.
ou have still not answered the questions:
What is the DIFFERENCE between Hebrews (Abraham, Isaac) and the JEWS (Ashkenazim, Falasha, Sephardim, etc.)?
Why if Hagar ‘the Egyptian’ was Arab and through matrillineal pedigree was Arab, why did this cause a split in the J1/J2 haplogroups?
If, as you say, the Falasha are descendant from J1/Jews why did the Y-DNA lineage change to E-M35 or E-M123?
Was Abraham’s brother by the same mother Y-DNA E?
Just how did the lineage change for the ‘Black Jews’?
Just how did the lineage change for the ‘Ashkenzi’?
Just how did the lineage change for the ‘European’ Jews?
How, if all are descended from Abraham?
The Twelve Tribes of Israel? You tell me.
The Parent of E-M35 is E-P2.
CN
No incursion. SUBSTANTIATES the EXTENT of Ethiopian ADMIXTURE. This is all the so-called admixture there is to be found in the population – none!. Showing us that there is no difference between the Y-DNA E-M35 in the Falasha and the general Ethiopian population. As goes the Y-DNA as Eastern Africa, so does the mtDNA L.
Dag.
NO ADMIXTURE!!!!!
CN
…get a dictionary!!!!!
You wrote…
“you are NOT comprehending what YOU copy/paste – QUOTE, “It is more likely, therefore, that the matching haplotype does not represent the incursion of Jewish ******maternal***** (emphasis mine: MATERNAL, NOT PATERNAL)lineages into the Ethiopian gene pool but that this haplotype instead substantiates the extent of Ethiopian *****admixture*****(empahasis mine – term means MIXED as in PATERNAL JEWISH LINEAGE)in the Falasha population.
INCURSION: a hostile entrance into or invasion of a place or territory, especially a sudden one; raid: The bandits made brief incursions on the village.
Try it again….
“It is more likely, therefore, that the matching haplotype does not represent the incursion of Jewish maternal lineages into the Ethiopian gene pool but that this haplotype instead substantiates the extent of Ethiopian admixture in the Falasha population.”
The Skeptic
ONE – you are NOT comprehending what YOU copy/paste – QUOTE, “It is more likely, therefore, that the matching haplotype does not represent the incursion of Jewish ******maternal***** (emphasis mine: MATERNAL, NOT PATERNAL)lineages into the Ethiopian gene pool but that this haplotype instead substantiates the extent of Ethiopian *****admixture*****(empahasis mine – term means MIXED as in PATERNAL JEWISH LINEAGE)in the Falasha population.
Do you EVEN read my answers – I clearly explained to you the difference between Hebrew and Jew. Although Abraham was a Semite(Hebrew), he was NOT, Jewish – that line started with Isaac.I NEVER said the Falsha are descended from the Jews – I said they are MIXED – as in Jewish Semitic White fathers, Black Ethiopian mothers.Some had BOTH Black Ethiopian parents, but were Jewish by religious conversion, HENCE, no Jewish line present. That explains the Falsha.
JEWS – 3 types – Ashkenazim, Mizhrahi,Sephardim,
Ashkhennazim – Jews born in Europe – especially Germany – 1-3% of their genetic material is European, heritage from the surrounding peoples they mixed with.
Mizrahi – Jews from the Arab world and adjacent, primarily Muslim-majority countries. This includes Jews from Iraq, Syria, Lebanon, Yemen, Iran, Afghanistan, Uzbekistan, Kurdish areas, the eastern Caucasus, India, and Ethiopia.
Sephardi Jews -Jews from Morocco, Algeria, or Turkey and the Iberian Peninsula – generally Spanish countries.
As for Abraham – Jews are descended from Abraham’s union with Sarah, his half sister. Arabs are descended from Abraham’s union with Hagar.
The Falsha are NOT descendants of Semitic lineage – they are MIXED peoples – Black Ethiopians with Jewish Semitic White – MIXED.
CN
From your source above, QUOTE,
”Consistent with Y-chromosomal findings, this fact points to extensive admixture of Jews with the local population.”
Wrong interpretation: Translation – on the Patrilineal heritage – Jewish men took Black Ethiopan women as wives, on the matrilinear, the reverse is NOT true – ONLY Black Ethiopian men taking Black Ethiopian wives – hence, the some of, BUT, not all the Falsha Black Ethiopian Jews are MIXED – some are by religious conversion – NOW, do you understand – bottom lime – some are MIXED, some are by religious conversion.
Right interpretation:
Y-Chromosomal findings = Y-DNA E-M35 – Eastern Africa.
mt-DNA Haplogroup L – Eastern Africa.
Who are the ‘Jews’ in Ethiopia? the Falasha.
Therefore, the Falasha, i.e., the ‘Black Jews’ have Y-DNA and mt-DNA that is consistent with the general population of Ethiopia who are Y-DNA E-P2-M35.
“The E-M215 derivative, E1b1b1 (E-M35) is defined by the M35 SNP. E-M35 includes individuals with the “ancestral state” (no known sub-clade forming mutations). These are referred to as E1b1b1* or E-M35*. As of 2011, there are seven known branches that have resulted from different mutations on M35:
M68, V257, M123, V6, M293, V42 and V92.
In order to show what is known of their relationships to E1b1b1 and other related clades, these are also currently referred to as E1b1b1a to E1b1b1g, respectively (see image).
The more frequently described sub-clades are E1b1b1a (especially its more well-known sub-clade E-M78) and E1b1b1b (especially it well-known sub-clade E-M81). Both are found in Mediterranean Iberia & West Asian peoples.
These two sub-clades represent the largest proportion of E1b1b. E1b1b1a is found over most of the range where E1b1b is found excluding Southern Africa. E1b1b1b is found mainly in the Maghreb. E1b1b1c is less common but widely scattered, with significant populations in specific parts of the Horn of Africa, the Levant, Arabia, Iberia, and Anatolia.
NEW: E1b1b1e is a fourth major sub-clade that has been found in parts of Eastern and Southern Africa, includes the majority of unique E1b1b1 lineages in sub-Saharan Africa (those that lack M78, M81, or M123 mutations).[8] Three smaller sub-clades are defined by mutations V6, V42 and V92 appear to be unique to the Horn of Africa region.
Problem we just read this: ” E1b1b1a is found over most of the range where E1b1b is found excluding Southern Africa.”
Now read the new findings:
No E1b1b1A in Southern Africa but E1b1b1e is in both Eastern and Southern Africa.
NEW: “E1b1b1E is a fourth major sub-clade that has been found in parts of Eastern and Southern Africa, includes the majority of unique E1b1b1 lineages in sub-Saharan Africa (those that lack M78, M81, or M123 mutations).[8] Three smaller sub-clades are defined by mutations V6, V42 and V92 appear to be unique to the Horn of Africa region.”
Within E-M35, there are striking parallels between two haplogroups, E-V68 and E-V257. Both contain a lineage which has been frequently observed in Africa (E-M78 and E-M81, respectively) and a group of undifferentiated chromosomes that are mostly found in southern Europe. An expansion of E-M35 carriers, possibly from the Middle East as proposed by other authors, and split into two branches separated by the geographic barrier of the Mediterranean Sea, would explain this geographic pattern. However, the absence of E-V68* and E-V257* in the Middle East makes a maritime spread between northern Africa and southern Europe a more plausible hypothesis.
—Trombetta (2011)
E-M123 in Jews. Looking beyond simple regional concentrations, E1b1b1c (E-M123) is also quite common among both Ashkenazi and Sephardic Jews, accounting for over 10% of all male lines.
[http://en.wikipedia.org/wiki/Haplogroup_E1b1b_(Y-DNA)#CITEREFSemino_et_al.2004]
“It is more likely, therefore, that the matching haplotype does not represent the incursion of Jewish maternal lineages into the Ethiopian gene pool but that this haplotype instead substantiates the extent of Ethiopian admixture in the Falasha population.
Can you interpret this correctly now?
If not, I cannot help you.
You have still not answered the questions:
What is the DIFFERENCE between Hebrews (Abraham, Isaac) and the JEWS (Ashkenazim, Falasha, Sephardim, etc.)?
Why if Hagar ‘the Egyptian’ was Arab and through matrillineal pedigree was Arab, why did this cause a split in the J1/J2 haplogroups?
If, as you say, the Falasha are descendant from J1/Jews why did the Y-DNA lineage change to E-M35 or E-M123?
Was Abraham’s brother by the same mother Y-DNA E?
Just how did the lineage change for the ‘Black Jews’?
Just how did the lineage change for the ‘Ashkenzi’?
Just how did the lineage change for the ‘European’ Jews?
How, if all are descended from Abraham?
The Twelve Tribes of Israel? You tell me.
The Parent of E-M35 is E-P2.
Trombetta, Beniamino; Cruciani, Fulvio; Sellitto, Daniele; Scozzari, Rosaria (2011), MacAulay, Vincent, ed., “A New Topology of the Human Y Chromosome Haplogroup E1b1 (E-P2) Revealed through the Use of Newly Characterized Binary Polymorphisms”, PLoS ONE 6 (1): e16073, doi:10.1371/journal.pone.0016073, PMC 3017091, PMID 21253605 ;http://www.plosone.org/article/info:doi/10.1371/journal.pone.0016073
Get it right this time?
“Cruciani et al. (2004): “Several observations point to eastern Africa as the homeland for haplogroup E3b—that is, it had
1) the highest number of different E3b clades (table 1),
(2) a high frequency of this haplogroup and a high microsatellite diversity, and, finally,
(3) the exclusive presence of the undifferentiated E3b* paragroup.” As mentioned above, “E3b” is the old name for E1b1b (E-M215). Semino et al. (2004): “This inference is further supported by the presence of additional Hg E lineal diversification and by the highest frequency of E-P2* and E-M35* in the same region.
The distribution of E-P2* appears limited to eastern African peoples.
The E-M35* lineage shows its highest frequency (19.2%) in the Ethiopian Oromo but with a wider distribution range than E-P2*.” For E1b1b (M-215) Cruciani et al. (2007) reduced their estimate to 22,400 from 25,600 in Cruciani et al. (2004), re-calibrating the same data.”
Can you hear me now?
the Skeptic
Nothing cotradictory there – simply stated – the J haplo group is 30,000 years od, at3,300 years, the Arabs and jews diverged from a common ancestor.
READ WHAT YOU COPIED/PASTED ON THE FALSHA(Black Ethiopian)Jews- do so slowly.
From your source above, QUOTE,”Consistent with Y-chromosomal findings, this fact points to extensive admixture of Jews with the local population.” Translation – on the Patrilinear heritage – Jewish men took Black Ethiopan women as wives, on the matrilinear, the reverse is NOT true – ONLY Black Ethiopian men taking Black Ethiopian wives – hence, the some of, BUT, not all the Falsha Black Ethiopian Jews are MIXED – some are by religious conversion – NOW, do you understand – bottom lime – some are MIXED, some are by religious conversion.
CN….
You kind of skipped over this part…
http://www.simpletoremember.com/articles/a/abraham-lineage/
“Based on the DNA of today’s Kohanim, the geneticists have dated their “Most Common Recent Ancestor” to 106 generations ago, approximately 3,300 years before the present. This is in agreement with the Torah’s written and oral tradition of the lifetime of Aaron, the original High Priest and founder of the Kohen lineage. Further genetic studies have found that the CMH-the Cohen Modal Haplotype-a haplotype of the MED (J) haplogroup-is not exclusive to Kohanim, and not unique to Jews. It is also found in significant percentages among other Middle Eastern populations, and to a lesser extent, among southern Mediterranean groups. A haplotype is a group of distinct DNA markers—neutral nucleotide mutations, which when found together indicate a lineage. These particular markers were discovered on the Y-Chromosome, which is passed from father to son, without change, thus establishing a paternal lineage pattern.”
3000 years before present is 1048 B.C.
The Y-DNA is settled for the Falasha, i.e., ‘The Black Jews”. Here is the mtDNA:
“A majority of mtDNA lineages of Ethiopian Jews (Falasha or Beta Israel) derive from African-specific clades L0–L5 (Shen et al. 2004), including exact matches with Ethiopians sampled in the present study.
Consistent with Y-chromosomal findings, this fact points to extensive admixture of Jews with the local population. A specific haplotype match in haplogroup (preHV)1—which is also widely spread in the Near East—between Ethiopian Jews and non-Jews is more problematic, because it is also possible that the non-Jews obtained the lineage from the Jews. This particular (preHV)1 haplotype, with a rare transversion at np 16305, (1) has not been detected, so far, among other Semitic populations of the Near East; whereas, (2) in Ethiopia, it occurs both among Cushitic and Semitic speakers; and, (3) in Ethiopian Jews, there are many sub-Saharan African lineages from haplogroups L0–L3.
It is more likely, therefore, that the matching haplotype does not represent the incursion of Jewish maternal lineages into the Ethiopian gene pool but that this haplotype instead substantiates the extent of Ethiopian admixture in the Falasha population.
Taken together, the influx of the elements of the Hebraic culture in the first centuries A.D. probably did not have a major impact on the genetic pool of Ethiopians, and the present-day Jews of Ethiopian descent probably assimilated genes from the local non-Jewish populations through conversion of the latter to Judaism.
The other two episodes of intrusion of Semitic influence, related to contacts with southern Arabia, are weakly supported by our data. This is because, among the haplogroup N lineages present in high frequency in the Tigrais and other Ethiopian ethnic groups, only a few revealed close relationships with equivalent lineages present in southern Arabia.”
This ends the debate that the Falasha, i.e., ‘The Black Jews’ are in ‘Black-Africans.’
Any Southeast Asian (Eurasian) haplogroups make all other ‘Africans’ Afro-Asiatic and not less ‘Black’.
Now, your response to my response…
Well, it does not answer the questions…
What is the DIFFERENCE between Hebrews (Abraham, Isaac) and the JEWS (Ashkenazim, Falasha, Sephardim, etc.)?
Why if Hagar ‘the Egyptian’ was Arab and through matrillineal pedigree was Arab, why did this cause a split in the J1/J2 haplogroups?
If, as you say, the Falasha are descendant from J1/Jews why did the Y-DNA lineage change to E-M35 or E-M123?
Was Abraham’s brother by the same mother Y-DNA E?
Just how did the lineage change for the ‘Black Jews’?
Just how did the lineage change for the ‘Ashkenzi’?
Just how did the lineage change for the ‘European’ Jews?
How, if all are descended from Abraham?
The Twelve Tribes of Israel? You tell me.
the Skeptic
I TRULEY wish you were comprehending what you are copying/pasting – your own data is saying the Falsha(Black Ethiopian Jews differ from the surrounding Black Ethiopians because of the presence of the Semitic markers – back up on our answers – I have posted it TWICE now – no need to post it again – you are not reading it – the link shows the Jewish haplomarkers appearing in the Black Ethiopian Jews are PATRILINEAR(translation – Jewish men taking Black Ethiopian females). Several times now, I jhave posted the ethnic haplomarkers – those eith the subclade(not posted) define ethnic groups – yes, the lay person’s term of race for ethnic groupings , can IDENTIFY race – AGAIN, and, AGAIN, and, AGAIN – you keep identifying African as Black African, when all your copying/pasting identifies Black African as SUB Saharan. African in context of your studies – involve humanoidsof the human species from which all ethnic groups are derived from, INCLUDING BLACK, – NOW, White, or, any other ethnic group, are NOT descended from Black people, BUT FROM A COMMON ANCESTOR, who had dark hair, skin, and eyes – a mutation changed this between 13,000, and, 15,000 years ago. Your troble comes because you do NOT want to accept Semitic peoples as Caucasian – Semitic peoples have all the Caucasian haplomarkers European Whites do, plus four more t5hat make us Semites, that NO OTHER ethnic group has.You keep wanting to include the Semitoc haplomarkers in Black Africans – when, it only appears in MI(XED peoples – most of YOUR data indicates this – some presents contradictory evidence that i t may have originated in Africa(NOT, necessarily BLACKS) – I have established for you, the orugin of the haplomarkers, there was mixing of Arab Semitic Whites, and, Blacks, the populations of Egypt demonstrate this(even BY YOUR DATA) – Arab Semitic White, the Ancient and Modern Egyptians, NOT Black Nubians. Arab Semitic White in Northern Egypt(3/4 of Egypt) to Blacks in South Egypt(actually Southwestern 1/4. I also established that if you take skeletal samples from Northern Egyptian mummies, you get Arab Semitic White data, you take it from Black Southwestern 1/, you get Black data, You comnstrue this data to mean the Ancient Egyptians were Black, when, in reality, they were Arab SemiticW@hite. On Hebrews and Jewish, this article explains it nicely:
Judaism
HOME
ARTICLES
AUDIO & VIDEO
BLOG
EVENTS
Abraham’s Chromosomes?
In search of the historical Abraham
According to the written and oral traditions of the three major religions of the Western world, Abraham was a real person who lived in the Middle East nearly 4,000 years ago. According to each respective tradition, he was the first of the Fathers of the Jewish people, fathered the Arab nations and Islam, and laid the conceptual basis for Christianity. Tradition relates that he may have influenced early Eastern religion, as well.
Abraham is the first to be called a Hebrew – Ivri—one who passes over from one side to the other. He received this title because he actually crossed over Euphrates River, in present day Iraq, as he traveled to the Promised Land at the call of God. Philosophically, he earned the distinction as a Hebrew for his clarity of truth, for at a time that the entire world was of one opinion, he was of another. He was born, according to the Talmud, into a world that had largely lost recognition of the one God—the Creator, Sustainer, and Supervisor of the universe. He recognized at an early age that there must be only one Creator and Prime Mover of all. It was not a popular opinion at the time, but he was a fighter for truth and freedom, and he placed his life on the line for his belief. In his lifetime, he continually faced and passed major tests of his strength of conviction and commitment to his vision of truth of the reality and unity of God.
The Jewish people regard Abraham as their original forefather, the father of Isaac, and the grandfather of Jacob. Abraham is also revered as the forefather of the Arab nations and Islam, as he was also the father of Ishmael, his son through Hagar, Sara’s Egyptian princess handmaiden. The Koran reports that Abraham and Ishmael raised the foundations of the Kaaba, the cube-shaped black stone structure in Mecca, Saudi Arabia, which is Islam’s holiest shrine. During the annual Haj pilgrimage, Moslems from all over the world circle the Kaaba, reinforcing the central role of Abraham and Ishmael in Islamic faith. Christianity, as well, regards Abraham as a Patriarch. He is the acknowledged father of monotheism, the progenitor of Western religion.
Can recent genetic research give some indication of the existence of the historical Abraham?
Recent genetic studies of the Jewish people clearly indicate that the roots of the Jewish nation can be traced to the Middle East. This research confirms the geographical origin of the core of every major Jewish Diaspora community. (See: “Jewish Genetics”)
Furthermore, the discovery of the “Cohen Gene”—the genetic signature shared by the majority of Kohanim—the Jewish priestly family worldwide, is an indication that this signature is that of the ancient Hebrews. (See: “The Cohen – DNA Connection”)
Based on the DNA of today’s Kohanim, the geneticists have dated their “Most Common Recent Ancestor” to 106 generations ago, approximately 3,300 years before the present. This is in agreement with the Torah’s written and oral tradition of the lifetime of Aaron, the original High Priest and founder of the Kohen lineage. Further genetic studies have found that the CMH-the Cohen Modal Haplotype-a haplotype of the MED (J) haplogroup-is not exclusive to Kohanim, and not unique to Jews. It is also found in significant percentages among other Middle Eastern populations, and to a lesser extent, among southern Mediterranean groups. A haplotype is a group of distinct DNA markers—neutral nucleotide mutations, which when found together indicate a lineage. These particular markers were discovered on the Y-Chromosome, which is passed from father to son, without change, thus establishing a paternal lineage pattern.
All of the above is scientific fact, which has only become known in recent years. Using these findings as a basis, perhaps we can speculate and consider some implications of the findings.
If the CMH is the genetic signature of Aaron, the father of the Kohanim, it must also have been the genetic signature of Aaron’s father, Amram, and that of his father, Kehat, and of his father, Levi. Levi’s father was Jacob who also must have had the CMH as his Y-Chromosome genetic signature, as did his father, Isaac.
Thus we arrive at Abraham. Abraham was only seven generations removed from Aaron, a matter of a few hundred years. Genetic signatures change slightly only over many generations. Thus, it is very reasonable to assume that the CMH, the most common haplotype among Jewish males, is therefore also the genetic signature of the Patriarch Abraham.
This would explain why we also find the CMH in high numbers among Arabs and other Middle Easterners today. These peoples traditionally claim to be the progeny of Abraham through his son Ishmael, who would also have to be carrying Abraham’s male genetic signature. These markers are also found among some southern Mediterranean and European peoples.
Besides the Jews, there are other populations that share the “Abrahamic Genetic Signature” as their primary Y-markers. These include Lebanese, Syrians, Druze, Iraqi Kurds, some southern and central Italians, and Hungarians. It is also found among some Armenians. These may be descendants of Abraham through his grandson Esau, brother of Jacob, some of whose progeny, according to Talmudic tradition, founded the early roots of the empire of Rome. As Isaac’s son and Abraham’s grandson, Esau would also have had these same Y-chromosome lineage markers. Please keep in mind that this part .>is the author’s speculation only.
The Jewish Kohanim have maintained the Abrahamic lineage to the highest degree among the Jewish People. Jewish is not a genetic definition—other peoples, through marriage and conversions, have joined the Jewish People. However, being a Kohen is a genetic definition—father to son starting from Aaron, the High Priest. And despite their having been scattered throughout the world for over 2,000 years, the extended family of Kohanim have maintained their genetic integrity equivalent to the highest percentages of the other Middle Eastern groups which never left the region.
. Based on the dating of the Most Recent Common Ancestor of the Kohanim as approximately 3300 years, it is not unreasonable to assume that it is the male descendants of Patriarch Abraham today who possess this DNA signature. However, Abraham may not be the exclusive source of these markers, for they are a component of a more ancient Middle Eastern gene pool.
“From Me, behold, I make My covenant with you, you shall be the father of many nations… And I will make you exceedingly fruitful, and I will make of you nations, and kings shall come from you.” Genesis 17:4, 6
The promise and prophecy of God to Abraham was that he would be the progenitor of great nations, that his descendents—literally “his seed”—would be numerous “as the stars in the heavens and as the sands on the seashore,” (Genesis 22:17). And indeed, the number of people in the world today with the “Abrahamic Genetic Signature” is too large to count precisely. A reasonable estimate is in the hundreds of millions.
This article is based on a chapter from the new book, DNA and Tradition: the Genetic Link to the Ancient Hebrews. Click here to order.: http://www.simpletoremember.com/articles/a/abraham-lineage/
by Rabbi Yaakov Kleiman
Posted in: Jewish History
What is the difference between ‘Black’ and ‘White’?
A gene : SLC24A5.
“The researchers then looked at two different human populations in which people with European and African ancestors had mixed relatively recently — African-Americans and African-Caribbeans. They found that, on average, people with two copies of the European version of the gene had the lightest skin. People with two copies of the non-European version of the gene had darker skin, and people with one copy of each version of the gene had skin color somewhere in between.”
The anthropology of race and the discovery of a skin color gene, SLC24A5 :
http://anthropology.net/2005/12/27/the-anthropology-of-race-and-the-discovery-of-a-skin-color-gene-slc24a5/
You know what this means don’t you. Genetics has just proven that the absence of color, ie, pale skin is a major evolutionary gene selection to fix the ‘races’.
Just saying…as I roll my eyes…being brown-skinned does not make me less ‘African’ – so stop with calling ALL haplogroups the product of ‘Caucasoid’ admixture because Africans are coded with the darker gene just as ‘Europeans’ gene for color is now fixed.
I suppose that is the most obvious difference between the ‘races’ while the nose and hair is less subjected to fixation in the diversity of African phenotypes, but ‘less subjected’ does not mean admixture with ‘Europeans’.
the Skeptic
The haploGroup J(Y-DNA) is 30,000-50,000 years old: http://en.wikipedia.org/wiki/Haplogroup_J_%28Y-DNA%29
Hagar, Abrahan’s second wife, was from Maghreb – Caucasoid,peoples – Abrahan was Hebrew, NOT, Jewish – lay people think they are one, and, the same. Abraham, however, was Semitic – hence, Jews and Arabs share the Semitic markers.AGAIN – you confuse African with Black African – NONE of your sources claim what YOU do – Black African equates with SUB – Saharan.
The presence of the J haplogroup from the Jewish Semitic White (doesn’t occur in Blacks – ONLY mixed – i.e -Mulattoes, and, Semitic White People)in the Falasha indicates some are mixed(part Jewish(probably some Arab to)/part Black Ethiopian). Try AGAIN friend – seems you do NOT want to accept the Arab – Jewish to- influence.
Listen, I don’t care if Abraham was Greek. My point was a daughter could have married an ‘African’ and started a new lineage of ‘Black Jews’.
Follow me here, dude because it is a question.
You say Ishamel was an ‘Arab’ and that obviously he was an ‘Egyptian’. His mother was Hagar, ‘the Egyptian’ who I guess was ‘Arab.’ So what caused the split between the sons of Abraham who was oh an Hebrew and not Jewish. So who then are the Jews?
Now that two questions?
Falasha: http://findarticles.com/p/articles/mi_qa3659/is_199912/ai_n8872400/
“The proportions of the 3 main haplotypes in Ethiopians (haplotypes V, VIII, and XI) are not much different from those recently reported by Passarino et al. (1998). Haplotype VI (A2,CO,D1,F0j1) is the third haplotype present in Falasha Jews (10.5%), but it is absent in our sample of Ethiopians. The distinctiveness of the Y-chromosome haplotype distribution of Falasha Jews from conventional Jewish populations (absence of haplotypes VH and VIII) and their relatively greater similarity in haplotype profile to nonJewish Ethiopians (presence of haplotypes V and Xl) are consistent with the view that the Falasha people descended from ancient inhabitants of Ethiopia who converted to Judaism (Ullendorff 1968). A similar genetic affinity of Falasha Jews with Ethiopians also emerges from analyses of the Y chromosome (Ritte et al. 1993a) and the nuclear and mitochondrial genomes (Hakim et al. 1990; Zoosmann-Diskin et al. 1991; Ritte et al. 1993b). Absence of haplotype VIII in Falasha Jews could be due to sampling error (only 38 Falasha Jews were examined in this study). The relatively high frequency of haplotype VI in the Falasha Jews-a haplotype rarely found in Europe (Lucotte and Hazout 1996) or in African populations (Spurdle and Jenkins 1992)-sets the Falasha Jews apart from Ethiopians. These singularities indicate a mode of genetic differentiation of Falasha Jews based on genetic drift, which is a major force on small isolated populations.”
ONE MORE TIME:
“DNA samples from Beta Israel/Falasha Jews and Ethiopians were studied with the Y-Chromosome-specific DNA probe p49a to screen for TapI restriction polymorphism and haplotypes. Two haplotypes (V and XI) are the most widespread in Beta Israel and Ethiopians, representing about 70% of the total number of haplotypes in Ethiopia. Because the Jewish Haplotypes VII and VIII are not represented in the Falasha population, we conclude that these people descended from ancient inhabitants of Ethiopia who converted to Judaism.
DNA & tradition: the genetic link to the ancient Hebrews By Yaakov Kleiman, p.83”
http://www.madote.com/2010/09/ethiopias-queen-of-sheba-and-ark-of.html
Black converts!
CN…just because it ‘looks’ like it to you does not make it true in genetics.
J1-M267 Y lineage marks climate-driven pre-historical human displacements
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2986692/?tool=pubmed
The present day distribution of Y chromosomes bearing the haplogroup J1 M267*G variant has been associated with different episodes of human demographic history, the main one being the diffusion of Islam since the Early Middle Ages. To better understand the modes and timing of J1 dispersals, we reconstructed the genealogical relationships among 282 M267*G chromosomes from 29 populations typed at 20 YSTRs and 6 SNPs.
Phylogenetic analyses depicted a new genetic background consistent with climate-driven demographic dynamics occurring during two key phases of human pre-history: (1) the spatial expansion of hunter gatherers in response to the end of the late Pleistocene cooling phases and (2) the displacement of groups of foragers/herders following the mid-Holocene rainfall retreats across the Sahara and Arabia.
Furthermore, J1 STR motifs previously used to trace Arab or Jewish ancestries were shown unsuitable as diagnostic markers for ethnicity…
“…in the Amhara from Ethiopia, we found the very first case of a M368(xM367) chromosome, which supports the insertion of the paragroup J1e1* in the latest Y haplogroup phylogeny.
“With the exception of the rare Palestinian modal haplotype, none of the previously described STR motifs resulted or equated by descent, as they were found across ethnic groups with different cultural or geographic affiliation and in other lineages (J2, I*) than J1.
Such results make their use to trace ancestries of individuals or communities (ie, Arab or Jewish) inconclusive. Calculations under the coalescent model for J1 haplotypes bearing the Cohanim motif gave time estimates that place the origin of this genealogy around 6.2 Kybp (95% CI: 4.5–8.6 Kybp), earlier than previously thought, and well before the origin of Judaism (David Kingdom, ∼2.0 Kybp.”
“As regards chromosomes bearing alleles Dys388*13 and YCAIIa*19, which are common in populations of the Eurasian pool and in Northern Caucasians, we, in general, obtained Late Pleistocene coalescence times – around 10.1–10.9 KyBP as average median values.
These time estimates, summed to the high variation and wide distribution of these subclades, are consistent with the episodes of spatial re-expansion that occurred after the Last Glacial Maximum in the northern hemisphere, the latest being triggered by the end of the Younger Dryas event (12.9–11.6 KyBP20).
The different pattern observed (namely, frequency peaks in the Caucasus and Anatolia for Dys388*13, and, in ETHIOPIA and SOUTH-WESTERN ASIA with the ABSENCE of HAPLOTYPES of the ‘ARABIC POOL’FOR YCAIIa*19) does not deviate from random expectations of frequency shifts under an extended Wright–Fisher model (P≫0.05).
So what looks like ‘Caucasian’ is Eurasian and they-Ethiopians are right back to being part of the ‘Afro-Asiatic’ history of African Semitic languages such as Amharic, Geeze, Ethiopic, Tigrina, Eritrean, etc. Modern Arabic is at the bottom of the Semitic Languages ladder.
SPECIAL MENTION:
I did find a mutation, 12af/12af2 together with a -(YAP) is correlated to ‘Caucasian’/Eastern Asian mutations [as opposed to South-Western Asian mutations in Ethiopians].
This gene (12fa2)is characterized as being ‘associated ‘ with Caucasoids but in terms of its being carried by Khoisan this was obviously part of gene encoding as an unexpressed phenotype. The deletion of 12fa2 suggests this ‘Caucasoid’ association. No deletion…it is rightly so an encoded gene subject to mutation by deletion, insertion, and, in some cases is obviously neutral!
Now, answer the question: You say that Ishmael, Abraham’s firstborn son by “Hagar the Egyptian’ was an Arab, so I assume this is matrillineal, since Abrham was not an Arab? Issac was An Hebrew – but not a Jew, like his father. So, how did they split into the ‘Hebrew’ BUT NOT JEWISH and ‘Arabic’ lineages?
And what is a Jew, since you say Abraham was an Hebrew?
I think everyone who reads these postings would like to know the DIFFERENCE BETWEEN JEWS AND HEBREWS?
There is nothing in this post that has anything to do with Africans, except how the Falasha [Y-DNA E-M35] became Jews if they are descended from Abraham [Y-DNA J] the Hebrew but not the Jew.
You wrote…
“Hagar, Abrahan’s second wife, was from Maghreb – Caucasoid,peoples – Abrahan was Hebrew, NOT, Jewish – lay people think they are one, and, the same. Abraham, however, was Semitic – hence, Jews and Arabs share the Semitic markers.AGAIN – you confuse African with Black African – NONE of your sources claim what YOU do – Black African equates with SUB – Saharan.
The presence of the J haplogroup from the Jewish Semitic White (doesn’t occur in Blacks – ONLY mixed – i.e -Mulattoes, and, Semitic White People)in the Falasha indicates some are mixed(part Jewish(probably some Arab to)/part Black Ethiopian). Try AGAIN friend – seems you do NOT want to accept the Arab – Jewish to- influence.
Abraham, if he were ‘Caucasian’ would have no historical claim to Egypt as his homeland, so why am I confused about ‘African’ versus ‘Black African’. Neither Hebrew or Arabs had anything to do with the founding of the Falasha Jews..or are they Hebrews?….mmm, how could he? They are Y-DNA E not Y-DNA J.
So are these Jews or Hebrews?
Y-DNA J:
“Approximately 30% to 40% of Jewish men are in the paternal line known as haplogroup J[Note 1] and its sub-haplogroups. This Haplogroup is particularly present in the Middle East, Southern Europe, and Northern Africa.[11]
Y-DNA E1b1b (M35):
Furthermore, 15 to 30% are in haplogroup E1b1b[Note 2](or E-M35) AND ITS SUB-haplogroups.”
Y-DNA R1a1a (R-M17):
“Two studies by Nebel et al. in 2001 and 2005, based on Y chromosome polymorphic markers, showed that Ashkenazi Jews are more closely related to other Jewish and Middle Eastern groups than to their host populations in Europe (defined in the using Eastern European, German, and French Rhine Valley populations).
However, 11.5% of male Ashkenazim were found to belong to R1a1a (R-M17), the dominant Y chromosome haplogroup in Eastern European populations. They hypothesized that these chromosomes could reflect low-level gene flow from surrounding Eastern European populations, or, alternatively, that both the Ashkenazi Jews with R1a1a (R-M17), and to a much greater extent Eastern European populations in general, might partly be descendants of Khazars. They concluded “However, if the R1a1a (R-M17) chromosomes in Ashkenazi Jews do indeed represent the vestiges of the mysterious Khazars then, according to our data, this contribution was limited to either a single founder or a few closely related men, and does not exceed ~12% of the present-day Ashkenazim.”.[16][17] This hypothesis is also supported by the D. Goldstein in his book Jacob’s legacy: A genetic view of Jewish history.[18] However, Faerman (2008) states that “External low-level gene flow of possible Eastern European origin has been shown in Ashkenazim but no evidence of a hypothetical Khazars’ contribution to the Ashkenazi gene pool has ever been found.”.[19]”
Lastly…
“Biologist Robert Pollack stated in 2003 that one cannot determine the biological “Jewishness” of an individual because “there are no DNA sequences common to all Jews and absent from all non-Jews”.[5]
A 2009 study was able to genetically identify individuals with full or partial Ashkenazi Jewish ancestry.”
So, were these Askhenazi J1, J2, R1b1a or E-M35?
Bottomline? Y-DNA J is Proto-Arab but already a lineage of multi-ethnic groups-speaking-Semitic languages with differing regional markers indicating distinct ancestry not the same ancestor.
Oh, and when you bring the Greeks from ..what 567 BC in the gene pool you are really not talking about ancient gene flow, and what Y-DNA J are the Greeks?
“A newer study by Semino et al. [10] has studied two samples of Greeks of size 84 and 59 (Macedonian Greeks). The focus was on two specific haplogroups E and J which are frequent in the Mediterranean region and can be used to detect population movements between Europe, Africa and the Near East. 2.4% of Greeks belong in haplogroup E-M123 and 21.4% in E-M78. Clades of E prevalent in Northern or Sub-Saharan Africa were not found. According to Cruciani et al. [11] most Greeks and other Balkan people belong to a SPECIFIC CLUSTER within haplogroup E-M78 that is found in lower frequencies outside the Balkans and marks migrations from the Balkan area. E-M123 and its daughter haplogroup E-M34 originated in the Near East in prehistoric times. As for haplogroup J, most Greeks (22.8% Greeks/14.3% Macedonian Greeks) belong to J-M172 and its subclades which is associated with Neolithic population movements. Only 1.8%/2.2% of Macedonian Greeks/Greeks belonged to haplogroup J-M267 which could potentially (although not certainly) reflect more recent Near Eastern admixture.”
Berbers:
“According to a recent study in 2011, in Tunisia, J1-M267 is significantly more abundant in the URBAN (31.3%) than in the RURAL TOTAL POPULATION (2.5%). According to the authors, these results could be explained supposing that Arabization in Tunisia was a military enterprise, therefore, mainly driven by men that displaced NATIVE Berbers to geographically marginal areas but that frequently married Berber women.[http://en.wikipedia.org/wiki/PubMed_Identifier ; http://www.nature.com/jhg/journal/vaop/ncurrent/full/jhg201192a.html%5D”
“E3b1-M78, E3b3-M123, and R1*-M173 – mark gene flow between Egypt and the Levant during the Upper Paleolithic and Mesolithic.”
http://hpgl.stanford.edu/publications/AJHG_2004_v74_p000-0130.pdf
the Skeptic
We keep missing each other – READ my answers.
the skeptic – what in my answer, confused you – Arabs and Jews are Semitic Whites, sharing up to 40% of the same genetic material – Arabs are NOT Jews – the Northern HALF of Sudan, Northerm 1/3 of Somalia, and, Northern 1/3 of Ethiopia, is Arab Semitic White – mixing of the two peoples gives the peoples there, the more Caucasian looks – AGAIN – ADMIXTURE – NOT, PHENOTYPICAL VARIATION. What is so hard for you to accept??? Maybe this makes it easier – take President Obama – he is HALF Indo European Aryan White, and, HALF Black -though he is NOT White, NOR, Black, he has MORE Caucasian looks – lighter skin, thin lips, more narrow , elevated nose – higher forhead, less prognathous(projecting). Many Afro Centrists want to call him Black – he is NOT, Nor, is he White – he is mixed – his attributes go BEYOND phenotypical variation. The late Michael Jackson’s transformation is an ARTIFICIAL transformation – he went from looking Black, to Mixed, to White.
Answer the question.
If Abraham was J2 and both Jews and Arabs are descendant of Abraham. What caused the split in the lineage since Ishmael according to you is ‘Arab’ and according to the Jews Isaac is Jewish? Ishmael, the ‘Arab’ marries Hagar, the “Egyptian” who according to is ‘Arab’. So, what caused the split between the so-called Arab and Jews descendant from Abraham?
That is the question. If two sons from the same father can be divided into Arabs and Jews then what makes you think there is not a branch that can be added to divide as Arab, Jew, and African? All that has to happen is for an ‘African’ E1b* to marry a ‘Jewish’ maiden and a new lineage has begun whose sons, i.e, Ashkenazi or otherwise will also be E1b*.
What cause the split?
the Skeptic
Now, I am rolling my eyes – you are just NOT comprehending your OWN copy/paste. Arabs and Jews are Semitic White – yes, Arabs are NOT Jews – that is correct. Both share the J1(older) and J2(younger) haplomarkers – Arabs have more of the J1, Jews more of the J2 – translation – Arabs are older than Jews – Biblically, Ishmael(Arabs) before Isaac(Jews). Since both are Semitic White, sharing much of the same genetic markers – hard to tell which spread through Blacks in Africa since both Semitic Whites – Arabs and Jews have a presence there – FACT – most of the tribes you mentioned, are Black/Semitic White mixes – presence of the J1 & 2 haplomarkers show this – NOT present in Blacks – only in mixed peoples, Arabs, Jews, and a few other Semitic peoples – most, now, extinct. YOUR DATA SHOWS THIS – also, yo are STILL confusing African with Black African – – all ethnicities are descended from a COMMON AFRICAN ANCESTOR, NOT, FROM BLACK AFRICANS.Once you understand your erroneus thinking – you will see why you keep spinning your wheels. Very sinply – African does NOT equal Black African(MOST often called SUB – Saharan). Non Black Africans are sometimes referred to as SUPRA Sahara, North Africans, Arabs, South, Southwest Asians, Near East( NOW, Middle East). Berbers, and Moors, are a MIXED people – MOSTLY Arab heritage, some Black ancestry(usually 8-15%), small amounts of Jewish Semitic, and, Indo European Aryan , Whites – yes, I know your answer, and, it is simply, NOT,true – phenotypical variation – simply, NOT true – it is ADMIXTURE.
Roll your eyes…you wrote this…
“Bottom line, as simply as I can translate the studies for you – you keep skimmminig over the SemiticWhite (Arabs and Jews) presence in Ethiopians, Somalis, Sudanese that give these peoples more Caucasian looks than attributed to phenotypical variation. You keep skimming over the bidirectional flow of genes to, and, fromm the ME – the studies also show the Semitic White presence in Greeks who are Indo European Aryan Whites. The point is, you do NOT what to credit the Arabs with building the Pyramids, NOR, their influence in Black Africa, as well as the reverse – all the studies both of us quoted, back what I have stated and, you keep denying – i.e. – Falsha Jews are only Black – I presented the study show Jewish Semitic White paternal lineage in them, hence the more caucasian looks(keeps popping up). I also showed most of the other groups you claimed wwere Black tribes, were in reality, mixed Nlack/Arab Semitic White peoples. Your problem comes because you can not/will not accept this, instead, attributing it to phenotypical variation, so, in some way, as to say whatever achievements made, where, totally by Black people – I have shown you this is WRONG as far as the Pyramids and Ancient and Modern Egyptians are concerned – THEY ARE NOT THE BLACK NUBIANS, but ARAB SEMITIC WHITE.”
I was referring to the ‘Black Jews’ of Ethiopia
The point was to show both the ‘Jewish’ and ‘Arabic’ admixture in East Africans and North Africans (Sudan, included) population as being recent admixture as in the last 700 years or so…I stated…this quote:
“E1b1b and E1b1b1 are quite common amongst Afro-Asiatic speakers. The linguistic group and carriers of E1b1b1 lineage have a high probability to have arisen and dispersed together from the region of origin of this language family.[12][13][14] Amongst populations with an Afro-Asiatic speaking history, a significant proportion of Jewish male lineages are E1b1b1 (E-M35).[15] Haplogroup E1b1b1, which accounts for approximately 18%[3] to 20%[16][17] of Ashkenazi and 8.6%[18] to 30%[3] of Sephardi Y-chromosomes, appears to be one of the major founding lineages of the Jewish population.”
You said ‘White-Semitic-Jewish”. I said, wrong because E1b is Eastern Africa in origins! Any so-called White-Semitic-Jews’ either have shared paternal link with ‘Black’ Africans or the SNPs are totally different between both populations!
You cannot MAKE Y-DNA E-M35 ‘White’ because it suits you!
“A large majority of E1b1b lineages are within E1b1b1 (defined by M35). Exceptions discovered so far are M215 positive/M35 negative (“E-M215*”) cases found in two Amharic Ethiopians and 1 Yemeni.[2][20] At least some of these men, perhaps all, are known since early 2011 to be in a rare sibling clade to E-M35, E-V16/E-M281 (E1b1b2).[21] The discovery of M281 was announced by Semino et al. (2002), who found it in two Ethiopian Oromo. Trombetta et al. (2011) found 5 more Ethiopian individuals and an equivalent SNP to M281, V16. It was in the 2011 paper that the family tree position was discovered as described above.
The E-M215 derivative, E1b1b1 (E-M35) is defined by the M35 SNP. E-M35 includes individuals with the “ancestral state” (no known sub-clade forming mutations). These are referred to as E1b1b1* or E-M35*. As of 2011, there are seven known branches that have resulted from different mutations on M35: M68, V257, M123, V6, M293, V42 and V92. In order to show what is known of their relationships to E1b1b1 and other related clades, these are also currently referred to as E1b1b1a to E1b1b1g, respectively (see image). The more frequently described sub-clades are E1b1b1a (especially its more well-known sub-clade E-M78) and E1b1b1b (especially it well-known sub-clade E-M81). Both are found in Mediterranean Iberia & West Asian peoples. These two sub-clades represent the largest proportion of E1b1b. E1b1b1a is found over most of the range where E1b1b is found excluding Southern Africa. E1b1b1b is found mainly in the Maghreb. E1b1b1c is less common but widely scattered, with significant populations in specific parts of the Horn of Africa, the Levant, Arabia, Iberia, and Anatolia.
…..E1b1b1e is a fourth major sub-clade that has been found in parts of Eastern and Southern Africa, includes the majority of unique E1b1b1 lineages in sub-Saharan Africa (those that lack M78, M81, or M123 mutations).[8] Three smaller sub-clades are defined by mutations V6, V42 and V92 appear to be unique to the Horn of Africa region.”
Nothing ‘White’. Nothing ‘White-Semitic’
As for the ‘Arab’ admixture of Arab-speaking Somali males…
—you threw in the Arab Semitic white…
E1b1a (SNP) M78: Qatar – 1.39, UAE – 0.61, Oman – 2.00, Somalia-77.60, Egypt – 18.00, Lebanon – 10.48, Jordan – 10.27, Iraq – 9.36, Anatolia – 4.97, Iraq – 3.33, Pakistan – 1.14.”
CN
Duh! I forgot this..
Re : Sub-Saharan Sickle Cell haplotypes and E* haplogroups not present in Greek populations of Y-DNA E:
“In Africa the Hb S gene is associated with at least three haplotypes representing independent mutations.14, 15.
…They are
the Benin haplotype,
the Senegal haplotype and
the Central African Republic or the Bantu haplotype found in the central west Africa, the African west coast and the Central Africa (Bantu speaking Africa) respectively.
…A fourth haplotype, the Asian haplotype is found in the eastern province of Saudi Arabia and central India.”
Available calculations suggest that this gene has developed between 3000 and 6000 generations, approximately 150,000 – 70,000 years ago.
Obviously the inheritance of this haplotype in African populations migrating to Asia, where it mutated to the less-extreme haplotype,and entered Central Asia during those THOUSANDS OF YEARS, BEFORE ‘Europeans’ fixed-color coded gene mutated to switch off from dark to fixed-pale.
CN…this is ONE 2009 study so unless geneticists have repeated this study with other samples of the same populations, the expectation is the frequencies could increase. Until then…
Y-DNA Haplogroup E (Variegatde SNPs) Table: Abu-Amero et al. BMC Genetics 2009 10:59 doi:10.1186/1471-2156-10-59
http://www.biomedcentral.com/1471-2156/10/59/table/T1
E* – “The most basal lineages, paragroup E*, have been found in a single Bantu-speaking male from South Africa, amongst pygmies and Bantus from the Cameroon/Gabon region, and in two individuals from Saudi Arabia.”
E* (SNP) M96: SAR – 1.27, Lebanon – 0.55.
E1 (SNP) P147, descendants : E1a (M33), E1b (P177 : Follow the migration of Haplogroup E1 (P147) from East Africa to Arabia: http://www.thegeneticatlas.com/E1b_Y-DNA.htm
E1a* – While there have been no attested exemplars of E1*, its sub-clade, E1a (M33), is found most often in West Africa, and today it is especially common in the region of Mali. One study has found haplogroup E1a-M33 Y-chromosomes in as AS MUCH AS 34% (15/44) of a SAMPLE of MALIAN men. Haplogroup E1a also has been detected among samples obtained from Guinea-Bissau, Moroccan Berbers, Burkina Faso, Northern Cameroon, Senegal, Sudan, Egypt, Calabria (including both Italian speakers at 1.3% and Albanian speakers at 2.9%), Trentino (1/67 or 1.5%), and PORTUGAL(5/553 Sample or approximately 0.9%).
E1a has been detected in North Africa and EUROPE INDEPENDENLY OF the ubiquitous E1b1a. Because E1b1a is known to have EXPANDED RECENTLY, this leaves open the possibility of an ancient EXPANSION FROM West Africa INTO North Africa and Europe of E1a lineages.
E1a* (SNP) M33: EGYPT – 1.00, Lebanon – 0.22, Mali – 34% (Sample 15/44); detected in Guinea-Bissau, Moroccan Berbers, Burkina Faso, Northern Cameroon, Senegal, Sudan, Calabria (including both Italian speakers at 1.3% and Albanian speakers at 2.9%), Trentino (1/67 or 1.5%), PORTUGAL(5/553)0.9%.
E1b1a (SNP) P2: Anatolia – 0.38.
E1b1a (SNP) M2: SAR – 3.82, Oman – 5.00, Yemen – 3.23, Somalia – 1.50, Egypt – 1.00, Jordan – 0.66.
E1b1a7 (SNP) M191: SAR – 3.82, Qatar – 2.78, UAE – 5.52, Oman – 2.00, Egypt – 1.00.
E1b1b is less common in West, Central, and Southern Africa, though it has been observed among Khoisan speakers and among Niger–Congo speakers in Senegambia, Guinea-Bissau, Burkina Faso, Ghana, Gabon, the Democratic Republic of the Congo, Rwanda, Namibia, and South Africa.”
E1b1b is at once the most common E subclade amongst East African Maasai, Somalis, Ethiopians, Eritreans and North African Sudanese, Egyptians, Berbers and Arabs. It is also common in WEST ASIA, FROM WHERE it SPREAD into the Balkans and the rest of Europe. E1b1b has at least four common subclades: E1b1b1a (E-V68), E1b1b1b (E-V257), E1b1b1c (E-M123), and E1b1b1e (E-M293), the last of which spreads from Ethiopia to South Africa.
“The modern population of E-M215 and E-M35 lineages are almost identical, and therefore by definition age estimates based on these two populations are also identical. E1b1b (E-M215) and its dominant sub-clade E1b1b1 (E-M35) are believed to have first appeared in East Africa about 22,400 years ago.”
“E1b1b and E1b1b1 are quite common amongst Afro-Asiatic speakers. The linguistic group and carriers of E1b1b1 lineage have a high probability to have arisen and dispersed together from the region of origin of this language family. Amongst populations with an Afro-Asiatic speaking history, a significant proportion of Jewish male lineages are E1b1b1 (E-M35). Haplogroup E1b1b1, which accounts for approximately 18% to 20% of Ashkenazi and 8.6% to 30% of Sephardi Y-chromosomes, appears to be one of the major founding lineages of the Jewish population.”
“Within E-M35, there are striking parallels between two haplogroups, E-V68 and E-V257. Both contain a lineage which has been frequently observed in Africa (E-M78 and E-M81, respectively) and a group of undifferentiated chromosomes that are mostly found in southern Europe. An expansion of E-M35 carriers, possibly from the Middle East as proposed by other authors, and split into two branches separated by the geographic barrier of the Mediterranean Sea, would explain this geographic pattern. However, the absence of E-V68* and E-V257* in the Middle East makes a maritime spread between northern Africa and southern Europe a more plausible hypothesis.” —Trombetta (2011)
E1b1b* (SNP) M215: Yemen – 1.61
E1b1b1* (SNP) M35 : SAR – 2.55, Yemen – 3.23, Somalia – 1.50, Egypt – 3.00, Lebanon – 0.22.
E1b1b1a (SNP) M78: Qatar – 1.39, UAE – 0.61, Oman – 2.00, Somalia 0- 77.60, Egypt – 18.00, Lebanon – 10.48, Jordan – 10.27, Iraq – 9.36, Anatolia – 4.97, Iraq – 3.33, Pakistan – 1.14.
“Based on genetic STR variance data, Cruciani et al. (2007) suggests that E1b1b1a1 originated in “Northeastern Africa”, which in their study refers specifically to Egypt and Libya, about 18,600 years ago (17,300 – 20,000 years ago).[Wikipedia Note 5]
Battaglia et al. (2008) describe Egypt as “a hub for the distribution of the various geographically localized M78-related sub-clades” and, based on archaeological data, they propose that the point of origin of E-M78 (as opposed to later dispersals from Egypt) may have been in a refugium which “existed on the border of present-day SUDAN and Egypt, near LAKE NUBIA, until the onset of a humid phase around 8,500 BC.
The northward-moving rainfall belts during this period could have also spurred a rapid migration of Mesolithic foragers northwards in Africa, the Levant and ultimately onwards to Asia Minor and Europe, where they each eventually differentiated into their regionally distinctive branches”. Towards the south, Hassan et al. (2008) also explain evidence that some subclades of E-M78, specifically E-V12 and E-22, “might have been brought to Sudan from North Africa after the progressive desertification of the Sahara around 6,000-8,000 years ago”.
E1b1b1b1b (E-M183). This clade is extremely dominant within E-M81. In fact, while Karafet et al. (2008) continues to describe this as a sub-clade of E-M81, and ISOGG defers to Karafet et al., all data seems to imply that it should actually be considered phylogenetically equivalent to M81.
E1b1b1b* (SNP) M81: UAE – 0.61, Somalia – 1.50,Egypt – 8.00, Lebanon – 1.2, Jordan – 2.74, Anatolia – 0.19.
I hope this ends the pretense about Ethiopians being admixed ‘White – Semitic – ‘Jews.’
This should end the pretense that White-Semitic-Arab- chromosomes are prevalent in Eastern African ethnic groups.
You know, there are so many SNPs and STRs mutations that have been shown to reasonably stable in many regions and populations and the rare ones tell us more about the social interaction between regions.
You can’t make Y-DNA E* anything. It is as ‘African’ as it was when it migrated through Arabia as it was when it back-migrated from the Near East.
the Skeptic
Bottom line, as simply as I can translate the studies for you – you keep skimmminig over the SemiticWhite (Arabs and Jews) presence in Ethiopians, Somalis, Sudanese that give these peoples more Caucasian looks than attributed to phenotypical variation. You keep skimming over the bidirectional flow of genes to, and, fromm the ME – the studies also show the Semitic White presence in Greeks who are Indo European Aryan Whites. The point is, you do NOT what to credit the Arabs with building the Pyramids, NOR, their influence in Black Africa, as well as the reverse – all the studies both of us quoted, back what I have stated and, you keep denying – i.e. – Falsha Jews are only Black – I presented the study show Jewish Semitic White paternal lineage in them, hence the more caucasian looks(keeps popping up). I also showed most of the other groups you claimed wwere Black tribes, were in reality, mixed Nlack/Arab Semitic White peoples. Your problem comes because you can not/will not accept this, instead, attributing it to phenotypical variation, so, in some way, as to say whatever achievements made, where, totally by Black people – I have shown you this is WRONG as far as the Pyramids and Ancient and Modern Egyptians are concerned – THEY ARE NOT THE BLACK NUBIANS, but ARAB SEMITIC WHITE.
CN
(Sorry – Part I of III):
Well that is the beauty of being raised to read – to compare and contrast – to learn that even the most seemingly insignificant details can lead to a true re-writing of history. I do not need you to interpret anything for me.
You can call out all of the so-called ‘Caucasian’ admixture you want but the truth is Y-DNA haplogroup E began in ‘Black’ Africa. They were already genetically coded in skin color, just like the Khoi-San who are almost Mongoloid – looking with light complexion. They are predominantly md-DNA L0 and L1. Y-DNA A, B, E. By the way, the so-called Bantu expansion of 5,000 YBP or 3,048 BC was from Cameroon to Sub-Saharan Africa.
CN You opined…
“The point is, you do NOT what to credit the Arabs with building the Pyramids, NOR, their influence in Black Africa, as well as the reverse – all the studies both of us quoted, back what I have stated and, you keep denying – i.e. – Falsha Jews are only Black – I presented the study show Jewish Semitic White paternal lineage in them, hence the more caucasian looks(keeps popping up…”
First of all the discussion was NOT about who BUILT the PYRAMIDS!!!!!
Second, You jump from Falasha to Jews to Arabs…
White-Semitic-Jews then White-Semitic-Arabs….
Which is it? Are Arabs Jews by another name..like a flower by any other name is a rose?
Let me help you out now that we have settled the question that the ‘Falasha Jews’ are the ‘ Black Jews’ and are Y-DNA E – NOT ADMIXED WHITE-SEMITIC-JEWS OR ARABS.
“Haplogroup J is defined by a DNA marker known as M304, and is a branch of haplogroup IJ-M429. Everyone who carries the M304 marker today descends from a common paternal ancestor who lived roughly 30,000 years ago in the Middle East. Members of haplogroup J-M304 are found throughout Eurasia, the Middle East, and North Africa. Haplogroup J diverges into two main subhaplogroups, J1-M267 and J2-M172, with J1-M267 typical of eastern African and Arabian groups and J2-M172 more dominant in southern European and southwest Asian (Indian) populations. To date, more than thirty subgroups of haplogroup J-M304 have been described.”
The other Jewish haplotype and haplogroup is Y-DNA J1 and J2. Here is the timing for both:
Origin of the Jews and the Arabs: Date of their Most Recent Common Ancestor is Written in their Y-Chromosomes – However, There Were Two of Them – By Anatole A. Klyosov:
http://www.google.com/url?sa=t&source=web&cd=7&ved=0CEgQFjAG&url=http%3A%2F%2Fprecedings.nature.com%2Fdocuments%2F4206%2Fversion%2F1%2Ffiles%2Fnpre20104206-1.pdf&rct=j&q=HAPLOTYPE%20VIII%20is%20the%20ancestral%20haplotype%20in%20Jews.&ei=3OJTTpi8MM-EsgLJosWJBw&usg=AFQjCNFGkvNoRpIFdca70H3jEU22FpcjRQ&cad=rja
“A pattern of Y-chromosomal mutations in 37 and 67 marker haplotypes of the Jews and the Arabs indicates that their most recent common ancestor in haplogroup J1 (subclade J1e*) and that (a different one) in haplogroup J2 (subclade J2a*) lived 4,300 +/- 500 years before present (YBP) and 4,175 +/- YBP, respectively, that is practically at the same time.
…Then a split between the Jewish and the Arabic lineages in both J1 and J2 haplogroups occurred, which is clearly visible on the respective haplotype trees.
CN (Part II of II)
http://www.google.com/url?sa=t&source=web&cd=7&ved=0CEgQFjAG&url=http%3A%2F%2Fprecedings.nature.com%2Fdocuments%2F4206%2Fversion%2F1%2Ffiles%2Fnpre20104206-1.pdf&rct=j&q=HAPLOTYPE%20VIII%20is%20the%20ancestral%20haplotype%20in%20Jews.&ei=3OJTTpi8MM-EsgLJosWJBw&usg=AFQjCNFGkvNoRpIFdca70H3jEU22FpcjRQ&cad=rja
The date show that a common ancestor of Cohanim (Jewish High Priests of haplogroup J1 lived 1,070 +/- 170 YBP, while a common ancestor of Cohanim in haplogroup J2 lived 3,300 +/- 400 YBP (or 1348 BC)!!!!!
Refer to the same 2009 unknown sample number frequency study of Hg Y DNA J1 and J2 distribution
Y-DNA Haplogroup E (Variegatde SNPs) Table: Abu-Amero et al. BMC Genetics 2009 10:59 doi:10.1186/1471-2156-10-59
http://www.biomedcentral.com/1471-2156/10/59/table/T1 :
…J1* – M267 : SAR – 40.13, Qatar – 58.33, Oman – 38, Yemen – 72.58, Egypt – 19, Lebanon – 20.09, Jordan – 30.62, Iraq – 31.03, Anatolia – 8.41, Iran – 11.33, Pakistan – 3.41.
…J1-M365 – Anatolia – 0.19.
…J1e1-M368 – Anatolia – 0.19.
…J1e2-M369 – Anatolia – 0.19
…J2*-M172 – Oman – 5, SOMALIA – 0.5, Egypt – 8, Jordan – 6.16, Iraq 26.6, Anatolia – 14.34.
…J2a*-M410 – UAE – 1.23, Yemen – 4.84, Iran – 4, Pakistan 8.52.
…J2a-DYS413 – SAR – 11.46, Qatar – 2.78, UAE – 4.29, Iran – 9.33.
…J2a1-M47 – SAR – 2.55, Qatar – 1.39, UAE – 1.84, Lebanon – 15.28, Iran 3.33.
…J2a2*-M67 – SAR – 0.64, Qatar – 1.39, UAE – 0.61, Oman – 1, Yemen 4.84, Egypt – 3, Lebanon – 7.75, Jordan – 4.79, Anatolia – 3.63, Iran 2, Pakistan – 1.14.
…J2a2a*-M92 – UAE – 1.23, Lebanon – 0.11, Anatolia – 2.68, Iran – 1.33.
…158 – Anatolia – 0.38.
…J2a9-M339 – Anatolia – 0.19.
…J2a10-M340 – Anatolia – 0.19.
…J2b*-M12 – SAR – 1.27, Qatar – 1.39, UAE – 1.23, Egypt – 1, Lebanon – 2.73, Jordan – 2.05, Anatolia – 0.76, Iran – 2.
…J2b2*-M241 – Qatar – 1.39, Anatolia – .096, Iran – 1.33, Pakitan – 2.27.
Wikipedia: http://en.wikipedia.org/wiki/Haplogroup_J_%28Y-DNA%29
Lebanon Sample Size: 951 Males. Hg J1 – 18.9, J2 – 29.4. Total J1/J2 out of 951 Males is 48.3 or 459 Males. El-Sibai et al. (2009)
Part III of II:
http://www.google.com/url?sa=t&source=web&cd=7&ved=0CEgQFjAG&url=http%3A%2F%2Fprecedings.nature.com%2Fdocuments%2F4206%2Fversion%2F1%2Ffiles%2Fnpre20104206-1.pdf&rct=j&q=HAPLOTYPE%20VIII%20is%20the%20ancestral%20haplotype%20in%20Jews.&ei=3OJTTpi8MM-EsgLJosWJBw&usg=AFQjCNFGkvNoRpIFdca70H3jEU22FpcjRQ&cad=rja
A few things:
:-D Expansion of mt-DNA L3e in Africa around 12,000kya.
Expansion of mt-DNA H1/H1v, H1w, H1x – around 10,000kya.
:-D R1-V88 back migration to West-Central-Africa around 9,500 – 5,600kya.
:-D Bantu Expansion 5,000kya.
:-D mt-DNA L3e2b-M4c-16519C transition, 3,000kya – South India.
Jewish-Arab Common Ancestor 2,048 BC:
:oops: “In other words, we see the two lineages derived from a common ancestor who lived around 4,300 YBP or 2,348 BC, one encompasses many Jews, and another mainly non-Jewish people, among them the Arabs.”
RECAP: J24ab, – 675 YBP, Arabs and Turks. J2a41b – 550 YBP. 450 YBP – J2a – Arabs. The Main branch – J2a>Jewish branch – J2a.
Figure .3. 67 marker 131 haplotype tree composed of the Arabic (haplotype numbers below 100) and Jewish (haplotype numbers 100 and higher) haplotypes of J2 haplogroup. All haplotypes were collected from sources indicated in the text. Some subclades are indicated on the graph, as well as some timespans to common ancestors for “young” branches. Branches of subclades J2a4b and J2a4b1 contain only Jewish haplotypes, with the respective times to their common ancestors of 1,200 +/- 210 and 700 +/- 150 YBP.
700+150 = 850YBP or 850-1952 = 1,102 CE/AD!!!!!!
“The younger,” Jewish only branch on the right-hand side in Fig. 2 had a common ancestor who lived 1,020 +/- 170 YBP or 932 CE (AD) !.
Again, this is within the margin of error with 1,125 +/- 160 YBP obtained for the 37 marker dataset. The data is quite reliable, and calcuations employing the 25, 37, and all 67 marker haplotypes in Fig. 2 gave 900 +/- 195, 1,150 +/- 140 YBP, respectively, to the lifetime of a common ancestor of the Jewish population (the “younger” branch on the right in Fig. 2), all of them having the “Cohen Modal Haplotype.”
Okay?
So how can you argue that Jews and non-Jews –here are the Arabs – when that common ancestor is indistinct in time for J1 and J2? If Arabs are non-Jews then what are the people from Sudan at 45%? Also non-Jews but the admixture is of the ‘Arab’ J2 and not of the Neolithic either but after even the Bantu Expansion.
J2 is nearly nearly coincident with the Le3b2-16519C transition back-migration from India, 3,000kya or what 1,048 BC if using YBP (1952!
Wikipedia:
“Haplogroup J1, defined by the 267 marker is most frequent in the Arabian Peninsula Yemen(76%),[13] Saudi (64%),[14] Qatar (58%),[15] and Dagestan (56%).[8] J1 is generally frequent amongst Arab Bedouins (62%.[16] It is also very common among others such as those of the southern Levant, i.e. Palestinians(38.4%), Ashkenazi Jews (20%),[17] North Sudan (45%), Khartoum (74%),[18] Algeria (35%),[17] Iraq (33%),[17] Tunisia (31%),[19] Syria (30%), Egypt (20%),[20] and the Sinai Peninsula. The frequency of Haplogroup J1 collapses suddenly at the borders of Arabic speaking countries with mainly non-Arabic speaking countries, such as Turkey (9%), Iran (11%).[1][9] It is also highly frequent among Jews, especially the Kohanim line (46%) [1] [21].”
Dag on I keeps sticking….Part I of III, Part II of III, and Part II of III.
This is just more of those nasty details that the devil revels in…
http://www.google.com/url?sa=t&source=web&cd=7&ved=0CEgQFjAG&url=http%3A%2F%2Fprecedings.nature.com%2Fdocuments%2F4206%2Fversion%2F1%2Ffiles%2Fnpre20104206-1.pdf&rct=j&q=HAPLOTYPE%20VIII%20is%20the%20ancestral%20haplotype%20in%20Jews.&ei=3OJTTpi8MM-EsgLJosWJBw&usg=AFQjCNFGkvNoRpIFdca70H3jEU22FpcjRQ&cad=rja
“The younger,” Jewish only branch on the right-hand side in Fig. 2 had a common ancestor who lived 1,020 +/- 170 YBP or 932 CE (AD) !.
Again, this is withis the margin of error with 1,125 +/- 160 YBP obtained for the 37 marker dataset. The data is quite reliable, and calcuations employing the 25, 37, and all 67 marker haplotypes in Fig. 2 gave 900 +/- 195, 1,150 +/- 140 YBP, respectively, to the lifetime of a common ancestor of the Jewish population (the “younger” branch on the right in Fig. 2), all of them having the “Cohen Modal Haplotype.”
:oops:
:oops: J2 lived 3,300 +/- 400 YBP (or 1348 BC)!!!!!
…J2a*-M410 – UAE – 1.23, Yemen – 4.84, Iran – 4, Pakistan 8.52.
“To sum this section uo, the most recent common ancestor of the Jews and Arabs of haplogroup J2a-M410* lived 4,175 +/- 510 years ago, and he had the “J2 Cohanim” signature in his haplotype. Again, it is rather “J2 Abraham Modal Haplotype.” From him a split occurred between the Jewish and the Arabic lineages in haplogroup J2. The split is clearly visible on the haplotype tree in the 37 and the 67 marker format (Figs. 3 and 4). Around 3300 YBP, in the middle of the 2nd millennium BC, a line of Cohanim, haplogroups J2a*, split from the lineage and continued with the initial ancestral haplotype. Around 2500 YBP, in times of the Babylonian and Assyrian exciles, the J2 Cohanim-Sephardim lineage split, and continued with the two mutations at DYS385a,b (13-18 compared to the initial 14-17) in the ancestral haplotype. The rest of J2 Cohanim continued with the same ancestral haplotype, and passed a population bottleneck (a genetic drift occurred) in the middle of the 1st millennium AD.”
4,175 + 510 = 4,685 – 1952 = 2,733 BC. 2500 YBP = 548 BC.
http://www.google.com/url?sa=t&source=web&cd=7&ved=0CEgQFjAG&url=http%3A%2F%2Fprecedings.nature.com%2Fdocuments%2F4206%2Fversion%2F1%2Ffiles%2Fnpre20104206-1.pdf&rct=j&q=HAPLOTYPE%20VIII%20is%20the%20ancestral%20haplotype%20in%20Jews.&ei=3OJTTpi8MM-EsgLJosWJBw&usg=AFQjCNFGkvNoRpIFdca70H3jEU22FpcjRQ&cad=rja
So, if you want to keep arguing that Northern Sudanese are admixed ‘Arabs’ then just accept what that timespan tells anyone who can read and count backward to 1,000 CE/AD. It happened during the slow but stead influx of Arabs into the Mediterranen, and obviously Egypt and North Sudan.
But, like I said, the discussion did not begin with my dis-belief that Arabs built the pyramids but that the Falasha Jews were converts to Judaism, irrespective of their ‘religious’ traditions which are rejected by the other Jews of Y-DNA J2.
It is curious though isn’t it, that the Palestians of Palestine, ( Akka), show out of 101 Males tested, that J1 – 39.2, and J2 – 18.6, for a total of J1/J2 – 57.8. El-Sibai et al. (2009).
Khartoum = J1 – 74% of an unknown sample number and J1 – 45% of an unknown sample number in North Sudan!
The highest ever reported concentration of J2 was 72% in Northeastern Georgia! (Unlnown sample number also).
It is curious that the Sudanese of Khartoum register higher than Palestinians!
So, if you want to convince the world that ‘White-Semitic-Arabs’ built the pyramids – hey knock yourself out.
Thanks for opening that door though…I just found a study about the skeletons of Naqada and Ballas… you will have to rethink that Nubians were not Egyptians mantra…
Still here is the question you have to answer…
If Abraham had two sons, Ishmael and Isaac. Why did a split happen in this genetic line since both were Y-DNA J1/J2 – that is Abraham was J1/J2.
The Hebrew Bible tells us Abraham had Ishmael by the ‘Egyptian’ bondwoman. If the Egyptians were ‘Arabs’ as you say then why would a son by an ‘Egyptian’ csuse a split in the J1/J2 lineage?
You know, I actually agree with. Details are a pain in the ….
CN
You started off wrong…
I asked, whether YOU KNEW how many sub-clades of Y-DNA E that have been identified?
Also, the false impression given in the studies you and I quote from, have the sample size of the men in the studies. The Y-DNA H*/1 study of the Tuareg of the Fellahen was a sample size of 100 and of that sample 61% tested for the African specific SNPs – not European.
Having qualified small samples sizes for all of these studies, there is this….
Haplogroup E originated in EAST AFRICA. THEREFORE, it is NOT JEWISH-WHITE-SEMITIC! Your attempt at lumping all ‘JEWS’ into one group goes against the details of the studies…your quote…
“In contrast, Ethiopian Jews (Beta Israel) and Indian Jews (Bene Israel and Cochini) cluster with neighbouring AUTOCHTHONOUS POPULATIONS in Ethiopia and western India, respectively, DESPITE a clear paternal link between the Bene Israel and the Levant.
NOTE: !!!!!
…These results cast light on the VARIAGATED GENETIC architecture of the Middle East, and trace the origins of most Jewish Diaspora communities to the Levant.”
Autochthonous : Originating where it is found; indigenous. Ethiopia. India. Near East. Different regional/population SNPs/STRs.
Follow the migration of Haplogroup E1b (P177) from East Africa to Arabia: http://www.thegeneticatlas.com/E1b_Y-DNA.htm
This clearly speaks to TWO HAPLOGROUPS [E-M123, E-M78 /M34], AND [J-M172/J-M267]:
“A newer study by Semino et al. [10] has studied two samples of Greeks of size 84 and 59 (Macedonian Greeks). The focus was on two specific haplogroups E and J which are frequent in the Mediterranean region and can be used to detect population movements BETWEEN Africa, the Near East, and EUROPE (In that order of migration).
2.4% OF GREEKS belong in haplogroup E-M123 and
21.4% in E-M78. Clades of E prevalent in Northern or Sub-Saharan Africa were not found. According to Cruciani et al. [11] most Greeks and other Balkan people belong to a SPECIFIC CLUSTER a within haplogroup E-M78 that is found in lower frequencies outside the Balkans and marks MIGRATIONS from the Balkan area. E-M123 and its daughter haplogroup E-M34 originated in the Near East in PREHISTORIC times. As for haplogroup J,
…most Greeks (22.8% Greeks/14.3% Macedonian Greeks) belong to J-M172 and its subclades which is associated with Neolithic population movements. Only 1.8%/2.2% of Macedonian Greeks/Greeks belonged to haplogroup J-M267 which could potentially (although not certainly) reflect more recent Near Eastern admixture.”
…This is consistent with the historic interaction of the ‘Jewish’-Ethiopians (as Eastern Africans at the Levant), Greeks (as Caucasians at the Levant), and Near Eastern Jewish populations (at the Levant).
…You cannot put all Ethiopians in the ‘Caucasian’ haplogroup because the frequency today is about 5.4%. This shows the migration of the Ethiopian to the Levant in the Neolithic along with every other population having settled in the neighborhood.
…The modern ‘Jewish’ – Ethiopian population (Falasha means STRANGER! the Biblical GER)was then scattered up the Nile, southward, before, during, and after the sack of Israel, which by the way was being defended by African Kings and Queens of Kush and not Libyan, Asiatic, Near Eastern Middle Eastern, ‘Arab’ Kings of Egypt! In fact, it was the Ethiopian Kingdom of Axum, that embraced Judeo-Christianity that sacked the African Temples of Amun in Dangeil – not ‘Europeans’.
“Clades of E prevalent in Northern or Sub-Saharan Africa were not found in Greeks.” Is that true?
Okay. It is true that the most basal clades of Sub-Saharan Africa were not found in Greeks.
“The most basal lineages, paragroup E*, have been found in a single Bantu-speaking male from South Africa, amongst pygmies and Bantus from the Cameroon/Gabon region, and in two individuals from Saudi Arabia..”
Sub-Saharan African Affinities with Natufians and East Africans [as opposed to Caucasian Affinities with East Africans and Egyptians]:
However, modern frequencies of the Benin-type-West African sickle cell traits have been found in Europe:
“A late Pleistocene–early Holocene northward migration (from Africa to the Levant and to Anatolia) of these populations has been hypothesized from skeletal data (Angel 1972, 1973; Brace et al. 2005) and from archeological data, as indicated by the probable Nile valley origin of the “Mesolithic” (epi-Paleolithic)Mushabi culture found in the Levant (Bar Yosef 1987). This migration finds some support in the presence in Mediterranean populations (Sicily, Greece, southern Turkey, etc.; Patrinos et al. 2001; Schiliro et al. 1990) of the Benin sickle cell
haplotype. This haplotype originated in West Africa and is probably associated with the spread of malaria to southern Europe through an eastern Mediterranean
route (Salares et al. 2004) following the expansion of both human and mosquito populations brought about by the advent of the Neolithic transition (Hume et al.
2003; Joy et al. 2003; Rich et al. 1998).
…This northward migration of northeastern African populations carrying sub-Saharan biological elements is concordant with the morphological homogeneity of the Natufian populations (Bocquentin 2003), which present morphological affinity with sub-Saharan
populations (Angel 1972; Brace et al. 2005). In addition, the Neolithic revolution was assumed to arise in the late Pleistocene Natufians and subsequently spread into Anatolia and Europe Bar-Yosef 2002), and the first Anatolian farmers, Neolithic to Bronze Age Mediterraneans and to some degree other NEOLITHIC –BRONZE Age EUROPEANS
show MORPHOLOGICAL AFFINITIES with the NATUFIANS (and indirectly with SUB-SAHARAN populations; Angel 1972; Brace et al. 2005), in concordance with a process of demic diffusion accompanying the extension of the Neolithic revolution.” (Cavalli-Sforza et al. 1994).
Therefore, to clarify the truth: “Clades of E prevalent in Northern or Sub-Saharan Africa were not found.” However, West African haplotype of Benin Sickle Cell have been found. The reason more ‘Europeans’ are not having children with the DISEASE is because only one chromosome was inherited in the genome pool of ‘Europeans’. Y chromosome and mtDNA studies do not show RECENT sub saharan markers in modern Southern Europeans anywhere near the degree of the Sickle Cell frequencies in ‘Africans’. Additionally, since this particular gene is a male inheritance this means African males and so-called ‘European’ women during the late Pleistocene – early Holocene migration of the Natufians. In other words, they were not ‘Caucasian-Europeans by any stretch of the imagination. Remember. ‘White’ people pride themselves on being a pure race. Color is an inherited trait that only mutated along with other mutations to produce the morphology of the so-called ‘European’ with the previously coded fixed dark color gene switching off.
This was the foundation of the study you offered:
“Although the Horn of Africa is considered a GEOGRAPHIC PART OF SUB-SAHARAN AFRICA, we have analysed the Somali population separately in order to be able to compare the results with previously published data from other African populations.”
So what is your point now about frequencies of Arab admixture? See next post.
the Skeptic
On the Black Falsha Jews – I DID provide the link SHOEING PATERNAL JEWISH SEMITIC WHITE GENETIC HERITAGE – QUOTE,”The falsha have an Ethnic paternal limk to the Jews – King Solomon and his men visited Ethiopia. QUOTE,”In contrast, Ethiopian Jews (Beta Israel) and Indian Jews (Bene Israel and Cochini) cluster with neighbouring autochthonous populations in Ethiopia and western India, respectively, despite a clear paternal link between the Bene Israel and the Levant. These results cast light on the variegated genetic architecture of the Middle East, and trace the origins of most Jewish Diaspora communities to the Levant.” – translation- Jewish men had Black Ethiopian wives: The genome-wide structure of the Jewish people: http://www.nature.com/nature/journal/v466/n7303/abs/nature09103.html “
the Skeptic
FIRST – all along – Berber (and Moor) base stock is mostly Arab, with 8-15% Black heritage, smaller amounts of Jewish Semitic, and Indo European Aryan White.
Second – spread of Arabs into Africa:High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males
Juan J Sanchez1, Charlotte Hallenberg1, Claus Børsting1, Alexis Hernandez2 and Niels Morling1
1Department of Forensic Genetics, Institute of Forensic Medicine, University of Copenhagen, Denmark
2Departamento de Canarias, Instituto Nacional de Toxicología, La Laguna, Tenerife, Spain
Correspondence: Dr JJ Sanchez, Department of Forensic Genetics, Institute of Forensic Medicine, University of Copenhagen, 11 Frederik V’s Vej, DK-2100 Copenhagen, Denmark. Tel: +45 35 32 62 87; Fax: +45 35 32 61 20; E-mail: [email protected]
Received 2 September 2004; Revised 12 January 2005; Accepted 21 January 2005; Published online 9 March 2005.
Top of page
Abstract
We genotyped 45 biallelic markers and 11 STR systems on the Y chromosome in 201 male Somalis. In addition, 65 sub-Saharan Western Africans, 59 Turks and 64 Iraqis were typed for the biallelic Y chromosome markers. In Somalis, 14 Y chromosome haplogroups were identified including E3b1 (77.6%) and K2 (10.4%). The haplogroup E3b1 with the rare DYS19-11 allele (also called the E3b1 cluster italic gamma) was found in 75.1% of male Somalis, and 70.6% of Somali Y chromosomes were E3b1, DYS19-11, DYS392-12, DYS437-14, DYS438-11 and DYS393-13. The haplotype diversity of eight Y-STRs (‘minimal haplotype’) was 0.9575 compared to an average of 0.9974 and 0.9996 in European and Asian populations. In sub-Saharan Western Africans, only four haplogroups were identified. The West African clade E3a was found in 89.2% of the samples and the haplogroup E3b1 was not observed. In Turks, 12 haplogroups were found including J2*(xJ2f2) (27.1%), R1b3*(xR1b3d, R1b3f) (20.3%), E3b3 and R1a1*(xR1a1b) (both 11.9%). In Iraqis, 12 haplogroups were identified including J2*(xJ2f2) (29.7%) and J*(xJ2) (26.6%). The data suggest that the male Somali population is a branch of the East African population – closely related to the Oromos in Ethiopia and North Kenya – with predominant E3b1 cluster italic gamma lineages that were introduced into the Somali population 4000–5000 years ago, and that the Somali male population has approximately 15% Y chromosomes from Eurasia and approximately 5% from sub-Saharan Africa.
Keywords:
Y chromosome, biallelic markers, SNPs, microsatellite loci, E3b1, Somali population
Top of page
Introduction
East Africans are more related to Eurasians than to other African populations.1, 2, 3 Investigations of Y chromosome markers have shown that the East African populations were not significantly affected by the east bound Bantu expansion that took place approximately 3500 years ago, while a significant contact to Arab and Middle East populations can be deduced from the present distribution of the Y chromosomes in these areas.4, 5 The Y chromosome haplogroup E3a is found at high frequencies in the sub-Saharan, Bantu-speaking populations but at low frequencies in East Africa, while Eurasian haplogroups like J and K are found at various frequencies in East Africa.3, 4, 6, 7, 8, 9 However, the majority of Y chromosomes found in populations in Egypt, Sudan, Ethiopia and Oromos in Somalia and North Kenya (Boranas) belong to haplogroup E3b1 defined by the Y chromosome marker M78.9, 10 A special branch of E3b1, cluster italic gamma, which was defined by the presence of the otherwise rare Y STR allele 11 in DYS19, was observed in high frequencies in small samples of male Boranas (Oromos) in North Kenya, Ethiopian Oromos and Somali males, while the E3b1 cluster italic gamma was found in low frequencies in non-Oromos from Ethiopia, Bantus from Kenya, North Egyptians10 and was almost absent in populations outside the Horn of Africa. Other clusters of haplogroup E3b1 (alpha, beta and delta) that are found in European, Arab, North and East African populations were not found in Oromos from North Kenya (Boranas) or Ethiopia, and found in only one of 23 Somali males.10
We typed a set of 45 biallelic markers and 11 STR systems on the Y chromosome in a large population of male Somali immigrants to Denmark in order to define their Y chromosome lineages in details. In addition, 65 sub-Saharan Western Africans, 59 Turks, and 64 Iraqis were typed for the biallelic Y chromosome markers. The results were compared to those obtained in other relevant populations.
Top of page
Material and methods
Samples and DNA purification
A total of 389 DNA samples from unrelated males (the numbers of individuals are given in parentheses) from Turkey (59), Iraq (64), Somalia (201) (all immigrants to Denmark) and 65 sub-Saharan Western Africans from Mali (38), Ghana (16), Mauritania (three), Guinea Conakry (two), Liberia (two), Cote d’Ivoire (one), Guinea – Bissau (one), Senegal (one) and Cameroon (one) (all immigrants to the Canary Islands) were typed in duplicates for 45 biallelic Y chromosome markers. Blood on FTA cards or Qiagen-purified DNA was used. The protocol was approved by the Danish ethical committee (Ref. KF-01-037/03).
Biallelic marker typing
The PCR amplification, the single base extension (SBE) reaction and the determination of the biallelic markers were performed as described previously.11 The markers P2, M22, M70, M75, M128, M168, M201, M207, M269 and M304 were also typed using singleplex PCR conditions.4, 11, 12 For the marker V6,10 we used the PCR primer sequences: V6F: 5′-CCTATAGAGTCCCTGTCCCTGA-3′, V6R: 5′-CTTGCTGCTGAGTGAGCTTCT-3′ (0.4 muM of each primer). SBE primers not described by Sanchez et al11 (0.2 muM of each primer) are given in Table 1.
Table 1 – Sequences of single base extension primers that have not been published previously.
Table 1 – Sequences of single base extension primers that have not been published previously – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the authorFull table
The haplogroup relationships and the nomenclature were established by the Y Chromosome Consortium (YCC).5, 13 The relevant portion of the YCC tree is shown in Figure 1.
Figure 1.
Figure 1 – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the author
Phylogenetic distribution of the 43 Y chromosome haplogroups that can be detected by the 45 biallelic markers. The arrow indicates the ancestral root of the maximal parsimonious YCC tree (2003).5 The major divisions of human Y chromosome diversity are labelled with large, capital letters in bold. On the right is shown the distribution of Y chromosome haplogroups in Somalis and in people from sub-Saharan West Africa, Turkey and Iraq. The relative frequencies in percent are shown in parentheses. aBecause none of our subjects studied belong to the haplogroup E3b1b, defined by the presence of M224,4 we used the haplogroup name E3b1 instead of E3b1*(xE3b1b) in the text.
Full figure and legend (57K)
STR typing
Y STR typing was performed using the PowerPlex® Y System kit (Promega) including DYS391, DYS389I, DYS389II, DYS439, DYS438, DYS437, DYS19, DYS392, DYS393, DYS390 and DYS385. The PCR conditions were as recommended by the manufacturer except that the number of cycles was 10 plus 16. The amplifications were performed in a Perkin–Elmer GeneAmp® PCR System 9700 thermal cycler. A total of 1 mul of PCR product was electrophoretically separated on an ABI 3100 Genetic Analyzer (Applied Biosystems) using performance-optimized polymer 4 (POP4) and Dye Set Z. Analyses of PCR fragments were performed using GeneScan 3.7 and Genotyper 3.7 NT (Applied Biosystems). The alleles were assigned by comparison to allelic ladders using Genotyper macros supplied with the PowerPlex® Y System. All 201 Somali males and the Turkish and Iraqi males previously assigned to belong to the haplogroup E3b1 were typed in duplicate. The nomenclature for DYS389II reflects the total number of repeats minus the number of repeats of DYS389I.14
As suggested by de Knijff,15 Y chromosomes identified by STRs are designated ‘haplotypes’. Y chromosomes that are defined only by biallelic markers are called ‘haplogroups or clades’ and the combination of biallelic markers and Y-STRs are called ‘lineages’.
Typing of 15 autosomal STRs was performed using the AmpFlSTR Identifiler PCR Amplification Kit (Applied Biosystems) according to the instructions of the manufacturer. A total of 198 of the 201 Somali males were typed in duplicate. The last three Somali males were not typed due to technical problems.
Comparative data analyses
In order to compare the proportions of Y-STR haplotypes in Somalis with those in other populations, we searched the worldwide Y-STR Haplotype Reference Database (http://www.yhrd.org/index.html). In addition, we compared the Somali Y STR data with results in African and Anatolian populations available in the literature. Phylogenetic relationships between Somali STR haplotypes within haplogroups were reconstructed in a median-joining network16 using the programme Network 4.1.0.0 (http://www.fluxus-technology.com). A haplogroup-specific weight proportional to the reciprocal of the microsatellite variance was used. Reduced-median and median-joining (alt epsilon=0) procedures were applied sequentially.
Statistical analyses
We used GENEPOP version 3.417 to test for Hardy–Weinberg proportions of the alleles of the 15 autosomal STR systems and calculations of the Fis values.18
The software package Arlequin, version 2.00019 (http://lgb.unige.ch/arlequin) was used to determine the molecular variance (AMOVA) at various levels of hierarchical groupings based on haplogroup frequencies20 and the mean pair wise differences, which are the mean number of mutational steps observed between all pairs of haplotypes in the sample.
The significance of the variance components and the corresponding Phi statistics (F statistics analogs) was assessed by comparisons of the observed values with the distribution of 10 000 permutations obtained by randomization under the null hypothesis of no population structure. The AMOVA was made considering the subpopulation relative to the total population (PhiST) and the geographical group of a subpopulation relative to the total population (PhiCT).6, 8 Principal component analysis of the haplogroup frequencies was performed using the Multivariate Statistical Package (MVSP) v. 3.1 (http://www.kovcomp.co.uk/mvsp/
mvspwbro.html) and presented graphically in two dimensions.
Y chromosome STR data were used to estimate the expansion time using both a model that does not explicitly specify a demography (through a star-like genealogy) and a Bayesian-based coalescence analysis procedure21 assuming a stepwise mutation model. The first approach required identification of the ancestral haplotype within each haplogroup using the DeltaA statistics described by Stumpf and Goldstein.22 To estimate the time to the most recent common ancestor (TMRCA), we used the average squared difference (ASD)23 and the averaged effective mutation rate described by Zhivotovsky et al.24 We used 95% confidence intervals (CIs) estimated by Monte Carlo simulations using a coalescent model with exponential growth scaled in units of the TMRCA including the uncertainty of the mutation rate. This is implemented in the programme Ytime described by Behar et al.25 Generation times of 25 and 30 years were assumed. Thus, the 95% CI takes into account the uncertainty in mutation rate, the population growth and (where appropriate) subdivision, but not the generation time. We also calculated the TMRCA using the variance of repeat scores observed (averaged over loci) within a haplogroup.26, 27
Bayesian analysis of trees with internal node generation (BATWING)28 was used to estimate the expansion times of a set of Somali Y chromosomes. The demographic model assumed exponential growth from an initially constant-sized population beginning at time Beta. Priors were chosen to be as uninformative as possible in order to minimize the impact on the results. Thus, we specified a gamma (1, 0.001) distribution as the prior for the growth rate and a gamma (1.1, 0.0001) distribution as the initial population size.12, 29 The prior distribution for the STR mutation rate was specified as a gamma distribution with a mean of 6.9 times 10-4 per locus per 25 or 30 years,24 and a broad, uniform Beta prior (0, 15) distribution was assigned. The estimated time of population expansion, Beta, was expressed as a fraction of the initial population size multiplied by the generation time to generate standard units of time. A total of 20 000 initial rearrangements were discarded, and the posterior distributions were estimated from the subsequent 50 000 rearrangements using the R computer software Version 1.8.1 (http://www.r-project.org/).
The results of microsatellite loci DYS385 were excluded from the statistical comparison analysis except for the network analysis because it was impossible to assign alleles to a specific locus. Estimation of the genetic diversity was measured using Nei’s unbiased h statistics.30 The variance of the allele distribution was calculated in EXCEL (Microsoft) for each locus independently and then averaged across the 10 loci.
Although the Horn of Africa is considered a geographic part of sub-Saharan Africa, we have analysed the Somali population separately in order to be able to compare the results with previously published data from other African populations.
Top of page
Results
Autosomal STR typing
The genotypes of the 15 autosomal STR systems in 198 Somalis were in Hardy–Weinberg equilibrium (chi2=31.37, df=30, P=0.40), and the Fis values ranged from -0.047 to 0.038.
Y chromosome haplogroup variation
We identified a total of 23 Y chromosome haplogroups in 389 males from Somalia, sub-Saharan West Africa, Turkey and Iraq. Figure 1 shows the genealogical relationship of the haplogroups and their frequencies.
In Somali males, 14 haplogroups were identified. The frequency of the clade E3b was 81.1%, including 77.6% of the haplogroup E3b1 defined by the M78 mutation. The Eurasian haplogroup K2 was found in 10.4%, and 3.0% of the Somali Y chromosomes belonged to the major clade J. Only 3.0% of the Somalis had the sub-Saharan African haplogroups A3, B and E3a*(xE3a4). Less than 2.0% of the Somalis belonged to the Northwest African E3b2 lineage. In the present study, no individual belonging to E3b* chromosomes carried the V6 mutation, which identifies a subset of chromosomes assigned to E3b* (E-M35*).10
Among the sub-Saharan Western Africans, only four haplogroups were identified. The West African clade E3a was found in 89.2%. Only one individual carried the major clade E3b (1.5%), and the haplogroup E3b1 was not observed.
In Turks, 12 haplogroups were found. The four haplogroups J2*(xJ2f2) (27.1%), R1b3*(xR1b3d, R1b3f) (20.3%), E3b3 and R1a1*(xR1a1b) (both 11.9%) were the most frequent ones.
In Iraqis, 12 haplogroups were identified. The haplogroup J2*(xJ2f2) was the most frequent one (29.7%) followed by J*(xJ2) (26.6%).
Geographic distribution of the Y chromosome haplogroup E3b1
To examine the relationship between the Y chromosome haplogroups in the Somali, other African and non-African populations, we compared our data with the data from literature (Table 2). The high frequency (77.6%) of haplogroup E3b1 was characteristic of male Somalis. The frequency of E3b1 was significantly lower in Ethiopian Oromos (35.9%), Ethiopian Amharas (22.9%), Egyptians (20.0%), Sudanese (17.5%), Kenyans (15.1%),10 Iraqis (6.3%), Northern Africans (6.1%), Southern Europeans (0.5–5.1%) and sub-Saharan populations (below 1%).
Table 2 – Frequencies of Y chromosome haplogroups among various populations.
Table 2 – Frequencies of Y chromosome haplogroups among various populations – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the authorFull table
We analysed the variation in the frequencies of E3b1 in 17 geographically defined populations using the AMOVA test.20 A total of 61.2% of the variance (significantly greater than zero, P<10-4) could be attributed to differences within populations. The population data were grouped into (1) sub-Saharan Africans (four populations), (2) North and East Africans (seven populations) and (3) non-Africans (six populations), and the Phi statistics were calculated. High degrees of both inter- and intragroup variabilities (PhiCT=0.31, P<10-4; PhiSC=0.19, P<10-4) were observed. In North and East African populations (PhiST=0.25, P<10-4), the variability was mainly due to the high frequency of the haplogroup E3b1 in the Somalis (77.6%) compared to those in Sudanese (17.5%) and Northern Africans (6.1%). When the Somali population data were removed from the North and Eastern African group, the PhiST value decreased from 0.25 to 0.16.
Figure 2 shows a principal component analysis of the haplogroups. We observed a similar pattern in a neighbour joining, unrooted tree (data not shown). The two principal components accounted for 79.4% of the genetic variance observed mainly due to differences in the frequencies of the clade E and clade BR*(xE) (first component) and the E3a and E3b lineages (second component). The first component separated the non-Africans and the sub-Saharan Western Africans characterized by high frequencies of the clade E3a (Figure 2, axis 1). The North and East Africans were separated from the rest of the sub-Saharan populations (Figure 2, axis 2) mainly by different frequencies of haplogroup E3b1. The position of the Ethiopian Oromos was close to that of the Somalis due to the relatively high frequencies of the haplogroups E3b*(xE3b1) and E3b1. The Ethiopian Amharas and the Egyptian population were positioned between the African and the non-African populations, primarily due to the high frequencies of the clade BR*(xE).
Figure 2.
Figure 2 – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the author
Principal component analysis of the relative frequencies of Y chromosome haplogroups in the populations reported in Table 2. The vectors express the relative weight of each haplogroup in the first and/or second axis. The positive or negative values indicate with which end of the axis the haplogroups are associated. Thus, the first principal component (axis 1) explained 52.8% of the total variance, mainly due to differences in the frequencies in clade E and clade BR*(xE). The second component (axis 2) explained 26.6% of the total variance, mainly due to the differences in the frequencies of the E3a and E3b lineages.
Full figure and legend (19K)
Y chromosome haplotype diversity
The allele and haplotype frequencies of 11 Y-STRs were estimated in the 201 male Somalis. Table 3 shows the Y-STR allele frequencies in all males, in haplogroup E3b1 and in non-E3b1 males. In eight of the 11 Y-STR systems, a predominant allele with a frequency above 0.75 was found. The frequencies of the predominant alleles ranged from 0.90 to 0.99 in E3b1 Y chromosomes.
Table 3 – Frequencies of Y chromosome microsatellite (STR) alleles in Somalis.
Table 3 – Frequencies of Y chromosome microsatellite (STR) alleles in Somalis – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the authorFull table
A comparison between the Y-STR allele frequencies in our and another sample of 104 Somali males typed for the eight common Y-STR loci37 showed that the distributions of Y-STRs were very similar in the two populations (Fst=0.0007; P=0.285).
In Somalis, 96 haplotypes were identified with the 11 Y-STRs. The haplotype diversity was 0.9726plusminus0.005. No microsatellite haplotype was found in more than one haplogroup. The haplotype diversity of the eight Y-STRs (minimal haplotype) was 0.9575plusminus0.007 compared to an average of 0.9974 and 0.9996 in European and Asian populations in the ''Y-STR Haplotype Reference Database' (http://www.yhrd.org/index.html), 0.9884 in the Mozambican population38 and 0.9838 in the Tunisian population.39
Haplogroup E3b1 with the rare allele 11 of DYS19 (E3b1 cluster italic gamma)10 was found in 75.1% of all Somali males, and 96.8% of the E3b1 Y chromosomes carried DYS19-11. The majority of the E3b1 DYS19-11 Y chromosomes were characterized by DYS392-12, DYS437-14, DYS438-11 (96.2%) and further by DYS393-13 (91.0%).
The average difference in numbers of repeat units between STR haplotypes typed for 10 Y-STRs was 0.7plusminus0.1, and the variance in the allele size distribution was 0.59 repeat units indicating that the Somali Y chromosome haplotypes were very closely related to each other. Figure 3a shows a median joining network analysis of the relatedness of the 64 Y STR haplotypes of the E3b1 haplogroup. The network displayed star-like features, and the four most frequent haplotypes, which accounted for 47% of the entire E3b1 cluster italic gamma lineage, occupied central positions in the network.
Figure 3.
Figure 3 – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the author
Networks of the Y chromosome STR data in haplogroup E3b1. (a) The network was constructed with the reduced-median procedure followed by the median-joining procedure based on data of 11 Y STRs in males belonging to haplogroup E3b1 in the present Somali population sample. The grey and black circles represent the Somali Y chromosomes with 12 and more than 12 repeats in the DYS19 microsatellite, respectively. Unless indicated by a number on the pie, the area of each circle is proportional to the number of individuals with the haplotype. The smallest circle corresponds to one Y chromosome. The lengths of the branches are proportional to the number of mutational steps. (b) The network was constructed with the reduced-median procedure followed by the median-joining procedure by combining our data with previously published data of DYS19, DYS389I, DYS390, DYS391 and DYS392 in individuals belonging to haplogroup E3b112, 40 (personal communication). The broken line delimits the lineages characterized by E3b1, DYS19-11 and DYS392-12 in Somalis and Ethiopians.
Full figure and legend (140K)
The network of the Figure 3b was constructed by combining our data with previously published data for individuals belonging to haplogroup E3b1 (12, 40 and personal communication). It displayed star-like features with a clear geographic structure. The main branch of the E3b1 cluster italic gamma lineages is located on a branch defined by DYS392-12 in the Ethiopian part of the network.
The Somali E3b1 haplotype data were compared with results from Anatolia,12 the only E3b1 data available for the same set of Y-STR loci. The two populations shared only one haplotype (DYS19-13/DYS389I-13/DYS389II-17/DYS390-23/DYS391-10/DYS392-11/DYS393-13/DYS439-12). The E3b1 cluster italic gamma lineages were not found in Turkish or Iraqi males.
In the 21 Somali males belonging to haplogroup K2, the Y-STR haplotypes were organized with a common ancestor into three branches with several mutation steps between the haplotypes (data not shown).
Estimates of expansion time and population size
Table 4 presents estimates of ages and expansion times of the Somali E3b1 cluster italic gamma and the K2 lineages based on Y-STR data using different estimation procedures. By defining the ancestral haplotype as that with the modal allele for each STR system and calculating the average squared distance41 as well as the variance between this haplotype and other variants, we estimated the time back to the most recent common ancestor (TMRCA). In Somalis, the TMRCA was estimated to be 4000–5000 years for the haplogroup E3b1 cluster italic gamma and 2100–2200 years for the haplogroup K2 assuming a generation time of 25 years. Calculations based on a Bayesian coalescence approach (BATWING expansion time) indicated that the growth of the E3b1 cluster italic gamma in the Somali population started 1200 years ago (Table 4) with an initial population size of 1037 individuals. A similar analysis of haplogroup K2 resulted in a calculated expansion time of approximately 3300 years in a small male population of 109 individuals. The results did not change significantly when different prior probability distributions were applied (data not shown).
Table 4 – Estimated relative ages and expansion times in thousands of years of the E3b1 cluster bold italic gamma DYS392-12 lineages and the haplogroup K2 in Somalis.
Table 4 – Estimated relative ages and expansion times in thousands of years of the E3b1 cluster [gamma] DYS392-12 lineages and the haplogroup K2 in Somalis – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the authorFull table
Top of page
Discussion
The present study demonstrates that male Somalis has the highest frequency of the haplogroup E3b1 (77.6%) observed in any population studied until now. The great majority of the Somali E3b1 Y chromosomes studied (96.8%) carried the otherwise rare allele 11 of the DYS19 STR locus and, thus, belonged to the cluster italic gamma defined by Cruciani et al.10 The E3b1 cluster italic gamma was previously reported in five of seven (71.4%) male Boranas (Oromos) from North Kenya, in 52.2% of Somali males, and in 32.0% Ethiopian males.10 The majority of the E3b1 Y chromosomes (91.0%) were further characterized by the DYS19-11, DYS392-12, DYS437-14, DYS438-11, and DYS393-13 alleles. The Eurasian clade FR had a frequency of 15.4% and the typical sub-Saharan haplogroups A, B, E3a*(xE3a4), E2 and E3b* were found in only 5% of Somali males.
The network of the E3b1 lineages in the present Somali population sample (Figure 3a) displayed star-like features and we observed a low Y STR haplotype diversity and a very limited spread in the sizes of the STR alleles (Table 3), suggesting a coherent, common, recent ancestry. The network of the E3b1 lineages of previously published data of East African populations and our data (Figure 3b) demonstrate that the E3b1 cluster italic gamma lineages of the present Somali population sample are part of the East African E3b1 lineages. E3b1 cluster italic gamma lineages were observed in low frequencies in Bantus from Kenya, North Egypt, Morocco and Niger10 (Figure 4). In the present study, haplogroup E3b1 was found in 6.3% of Iraqis and none of them belonged to cluster italic gamma. Only 11 subjects with a DYS19-11/DYS392-12 pattern were reported outside the Horn of Africa in 26 654 subjects analysed in a worldwide set of 236 populations by November 2004 (http://www.yhrd.org/index.html). Taken together, the data suggest that the E3b1 cluster italic gamma DYS392-12 lineage was expanded in the Somali population.
Figure 4.
Figure 4 – Unfortunately we are unable to provide accessible alternative text for this. If you require assistance to access this image, please contact [email protected] or the author
Map of African areas where E3b1 cluster italic gamma has been observed (the numbers of individuals are given in parentheses).10 (1) Moroccan Arabs (54), (2) Northern Egyptians (21), (3) Ethiopian Jews (22), (4) Ethiopian Amharas (34), (5) Ethiopian Wolaytas (12), (6) Mixed Ethiopians (12), (7) Ethiopian Oromos (25), (8) Somalia (224 including our Somali data), (9) Boranas (Oromos) from Kenya (seven), (10) Bantus from Kenya (28), (11) Tuaregs from Niger (22). The haplogroups or remaining paragroups are represented by different fill patterns. Lineages excluded from a haplogroup are listed within parentheses after the name of the haplogroup. The distribution of the Cushitic language in East Africa is shown in grey.
Full figure and legend (69K)
Cruciani et al10 suggested that the E3b1 cluster italic gamma lineages originated in East Africa and estimated that the TMRCA was approximately 9600 years. We estimated that the E3b1 cluster italic gamma DYS392-12 lineages of the present Somali population sample originated 4000–5000 years ago, and that the expansion of the E3b1 cluster italic gamma DYS392-12 lineages in these Somalis involved a relatively small number of Y chromosomes (around 1000 males).
The time of the eastbound Bantu expansion was estimated to be 3400plusminus1100 years ago.24 Bantu populations have high frequencies of E3a haplogroups.4 We have observed only a few individuals with the E3a haplogroup in our Somali population, thus, supporting the view that the Bantu migration did not reach Somalia.42 It has been suggested that a barrier against gene flow exist in the region.43 The barrier seems to be the Cushitic languages and cultures to which Somalis belongs. The Cushitic languages belong to the Afro-Asiatic languages that are spoken in Northern and Eastern Africa. The Cushitic languages and cultures are mainly found in the Somalis and the Oromos, one of the two main groups inhabiting Ethiopia.44, 45, 46. The Somali and Oromo languages have a high degree of similarity and the two populations share many cultural characteristics. The Somali and Oromo people live in clans with special patterns of marriage and the Somali and Oromo people have complex, interwoven pedigrees.44, 45
The very high frequency of the E3b1 cluster italic gamma in our Somali population sample could be due to ascertainment bias or special clan or family relationships in the present sample of Somali immigrants to Denmark. No reliable information on geographic origin or clan relationship in the present Somali population sample was available. However, the genotypes of the autosomal STR systems were in Hardy–Weinberg equilibrium, indicating random mating in at least the last generation, and the distribution of Y-STR haplotypes in our Somali population was similar to that in a sample of Somali immigrants to Norway,37 indicating that these two population samples came from a larger, homogenous population of Somalis.
The haplogroup K2 was found in 10.4% of Somali males. Haplogroup K2 was suggested to have arisen in Eurasia.4, 9 K2 has a patchy distribution in Cameroon (18.0%), Egypt (8.2%), Ethiopia (4.8%), Tanzania (3.8%) and Morocco (3.6%), probably due to back migration.3, 7, 8, 9 Luis et al9 estimated an expansion time of 13.7–17.5 ky for the K2 lineages in Egypt. The BATWING expansion time estimated for K2 in our Somali population (3.3 ky) is consistent with an African southward dissemination of the K2 haplogroup. The observation of two Somali males with the M17 mutation (haplogroup R1a1*(xR1a1b)) may indicate a recent gene flow by migration from Eurasia.47, 48 A possible explanation is offered by the fact that from the 7th century onward, immigrant Muslim Arabs and Persians established trading posts along the Somali cost51, although also British, French and Italian people were present in Somalia in the region in the 19th and 20th century.
The distribution of the haplogroups J2*(xJ2f2) (0.5%) and J*(xJ2) (2.5%) in Somalis support the recent gene flow hypothesis. Haplogroup J*(xJ2) was probably spread by the Arab people.40 The ratio between the haplogroups J2/J*(xJ2) may be an indicator of the genetic components from populations like (1) Balkans, Turks, Georgians and Muslim Kurds and (2) Bedouin and Palestinian Arabs, respectively.40, 52 The ratio was 0.26 in the Oman population.9 The J2/J*(xJ2) ratio of 0.2 in the present Somali sample suggest a predominant gene flow of Arab Y chromosomes.
In conclusion, the data suggest that the male Somali population is a branch of the East African population – closely related to the Oromos in Ethiopia and North Kenya (Boranas) – with predominant E3b1 cluster italic gamma DYS392-12 lineages that probably were introduced into the Somali population 4000–5000 years ago, approximately 15% Y chromosomes from Eurasia and approximately 5% from sub-Saharan Africa. Work is in progress in order to study closely related populations with new informative markers to obtain a better understanding of the E3b1 lineages settlement process in East Africa.:http://www.nature.com/ejhg/journal/v13/n7/full/5201390a.html
Ancient differentiation…not predominately admixture!
“…These findings are not easily explained. One possible scenario is that an ancient differentiation of the E-P2 haplogroup occurred in loco (East Africa). However, this also implies a low mutability of the associated microsatellite motif (DYS392-12/DYS19-11). Alternatively, the microsatellite motif may be due to HOMOPLASY.”
:oops: (DYS392-12/DYS19-11)
These STR values for 17 markers visible in the video are as follows:
:oops: DYS 19 – 14 (? not clear)
DYS 385a – 11
DYS 385b – 14
DYS 389i – 13
DYS 389ii – 30
DYS 390 – 24
DYS 391 – 11
:oops: DYS 392 – 13
DYS 393 – 13
DYS 437 – 14 (? not clear)
DYS 438 – 12
DYS 439 – 10
DYS 448 – 19
DYS 456 – 15
DYS 458 – 16
DYS 635 – 23
YGATAH4 – 11
ISOGG 2011 Y-DNA Haplogroup E and its Subclades. Last revision date for this specific page: 6 August 2011.
http://www.isogg.org/tree/ISOGG_HapgrpE.html
The dreaded Wikipedia Source for haplotype V:
E1b1b1a1 (E-M78) is a commonly occurring sub-clade, widely distributed in North Africa, the Horn of Africa, West Asia, (the Middle East and Near East) “up to Southern Asia”,[1] and all of Europe.[22] The European distribution has a frequency peak centered in parts of the Balkans (up to almost 50% in some areas)[3][23] and Sicily, and declining frequencies evident toward western, central, and northeastern Europe.
Based on genetic STR variance data, Cruciani et al. (2007) suggests that E1b1b1a1 originated in “Northeastern Africa”, which in their study refers specifically to Egypt and Libya.[Note 4] about 18,600 years ago (17,300 – 20,000 years ago).[Note 5]
…Battaglia et al. (2008) describe Egypt as “a hub for the distribution of the various geographically localized M78-related sub-clades” and, based on archaeological data, they propose that the point of origin of E-M78 (as opposed to later dispersals from Egypt) may have been in a refugium which “existed on the border of present-day Sudan and Egypt, near Lake Nubia, until the onset of a humid phase around 8500 BC.
…The northward-moving rainfall belts during this period could have also spurred a rapid migration of Mesolithic foragers northwards in Africa, the Levant and ultimately onwards to Asia Minor and Europe, where they each eventually differentiated into their regionally distinctive branches”.
…Towards the south, Hassan et al. (2008) also explain evidence that some subclades of E-M78, specifically E-V12 and E-22, “might have been brought to Sudan from North Africa after the progressive desertification of the Sahara around 6,000-8,000 years ago”.
Sub Clades of E1b1b1a1 (E-M78)
There are four recognized sub-clades, which were mostly defined by Cruciani et al. (2006).
E1b1b1a1a (E-V12). Found in Egypt, Sudan, and other places. Has an important sub-clade E1b1b1a1a2 (E-V32) which is very common amongst Ethiopian Oromo, Borana Oromo from Kenya and Somalis.
E1b1b1a1b (E-V13). This is the most common type of E1b1b found in Europe and is especially common in the Balkans.
E1b1b1a1c (E-V22). Found in Egypt, the Middle East and other places.
E1b1b1a1d (E-V65). Associated with the Maghreb, but also found in Italy and Spain.
E1b1b1a1e (E-M521). Found in two individuals in Greece by Battaglia et al. (2008)
E1b1b1b (E-L19/V257)- Ethiopia – Trombetta et al. (2011)
Main article: E1b1b1b
“E1b1b1b is dominated by its dominant sub-clade E1b1b1b1 (E-M81) which was discovered first, and has been more discussed in published literature. V257’s discovery was announced in Trombetta et al. (2011).
…The authors felt that it showed a parallel with its sibling clade E-V68 (above) in the way that both clades show signs of having MIGRATED FROM AFRICA to SOUTHWESTERN EUROPE across the Mediterranean sea.
…They found 6 “E-V257*” individuals in their samples who were E-V257, but not E-M81. A Borana from Kenya, a Marrakesh Berber, a Corsican, a Sardinian, a southern Spaniard and a Cantabrian.
E1b1b1b1 (E-M81), formerly E1b1b1b, E3b1b, and E3b2, is the most common Y chromosome haplogroup in the Maghreb, dominated by its sub-clade E-M183. It is thought to have originated in the area of North Africa 5,600 years ago.[2][24] This haplogroup reaches a mean frequency of 42% in North Africa, decreasing in frequency from approximately 80% or more in some Moroccan Berber populations, including Saharawis, to approximately 10% to the east of this range in Egypt.[24][25][26] Because of its prevalence among these groups and also others such as Mozabite, Middle Atlas, Kabyle and other Berber groups, it is sometimes referred to as a genetic “Berber marker”.
…Pereira et al. (2010) report high levels amongst Tuareg in two Saharan populations – 77.8% near Gorom-Gorom, in Burkina Faso, and 81.8% from Gosi in Mali. There was a much lower frequency of 11.1% in the vicinity of Tanut in the Republic of Niger.
E-M81 is also quite common among North African Arabic-speaking groups. It is generally found at frequencies around 45% in coastal cities of the Maghreb (Oran, Tunis, Tizi Ouzou, Algiers).[27]”
Different haplotypes,genotypes, phenotypes, and haplogroups due to homoplasy.
Whether you like it or not ‘Berber’ does not mean a specific racial/ethnic phenotype as these were a multi-ethnic group linked by language and culture. Marriage or what passed for marriage links the different ethnic groups by blood lines, likely matrilineal.
A matriline is a line of descent from a female ancestor to a descendant (of either sex) in which the individuals in all intervening generations are female. In a matrilineal descent system (uterine descent), an individual is considered to belong to the same descent group as his or her mother. This is in contrast to the more currently common pattern of patrilineal descent.
Mt-DNA ‘Berbers’- RECENT STUDY! :
http://knol.google.com/k/mainstream-academic-research/2010-berber-mtdna-study-finds-berber/3q8x30897t2cs/44#
EXCERPTED TEXT
Ancient Local Evolution of African mtDNA Haplogroups in Tunisian Berber Populations
Frigi et al.
Human Biology (August 2010 (82:4)
Abstract
“Our objective is to highlight the age of sub-Saharan gene flows in North Africa and particularly in Tunisia. Therefore we analyzed in a broad phylogeographic context sub-Saharan mtDNA haplogroups of Tunisian Berber populations considered representative of ancient settlement.
…More than 2,000 sequences were collected from the literature, and networks were constructed.
…The results show that the most ancient haplogroup is L3*, which would have been introduced to North Africa from eastern sub-Saharan populations around 20,000 years ago.
…Our results also point to a less ancient western sub-Saharan gene flow to Tunisia, including haplogroups L2a and L3b. This conclusion points to an ancient African gene flow to Tunisia before 20,000 years BP.
…These findings parallel the more recent findings of both archaeology and linguistics on the prehistory of Africa. The present work suggests that sub-Saharan contributions to North Africa have experienced several complex population processes after the occupation of the region by anatomically modern humans.
…Our results reveal that Berber speakers have a FOUNDATIONAL BIOGEOGRAPHIC ROOT in AFRICA and that deep African lineages have continued to evolve in supra-Saharan Africa.”
So, even the Tuareg, whose Y-DNA is various including E-M81/M183, the consistent mt-DNA Macro-haplogroup L is dominant, even where mt-DNA H1 (61% – Tuareg)and mt-DNA U6 are detected.
Haplotype V = Haplogroup E3b – Highest Genome Diversity in Africa
The Study of Racialism
http://www.pubmedcentral.gov/articleren … d=15069642
“Hg E (fig. 1A) is observed in Africa, Europe, and the Near East and includes the subhaplogroups E-M33, E-M75, and the most widespread subclade, E-P2.
…The latter includes three clusters, two of which, E-M2 and E-M35, are the most widespread.
…Haplogroups E-M33 (fig. 1B), E-M75 (fig. 1C), and the not-shown E-P2* and E-M2 are virtually absent in European populations and appear to be geographically restricted to sub-Saharan Africa.
…The E-P2* lineages were observed mainly in Ethiopians, whereas E-M2, which is considered a signature of the Bantu expansion (Hammer et al. 1998; Passarino et al. 1998; Scozzari et al. 1999), shows its highest frequency (>80%) in Senegal and has been sporadically observed in North Africa and Iraq.
…E-M35 (fig. 1D) has been found in Africa, the Near East, and Europe, where it is believed to have arrived in Neolithic times (Hammer et al. 1998; Semino et al. 2000).
…In particular, from among its subgroups, E-M78 (fig. 1E) is present in Europe, the Middle East, and North and East Africa. However, whereas no preferential YCAII microsatellite motif is observed in the Middle East, prevalent associations with YCAIIa21-YCAIIb19 in Europe and YCAIIa22-YCAIIb19 in Africa are found.
…E-M81 (fig. 1F) is almost absent in Europe (with the exception of Sicily and Iberia) and the Middle East but characterizes the majority of the Y chromosomes of populations from northwestern Africa.
…E-M123 (fig. 1G) is spread in the Near East and is also observed in North Africa and Europe but does not reach the western European regions.
…E-M281 and E-M329 are geographically restricted, having been seen only in Ethiopians (two subjects each).
…The remaining 37 E-M35* Y chromosomes were found mainly in Africa, with a high frequency in the Ethiopians and the Khoisan.”
Both phylogeography and microsatellite variance suggest that E-P2 and its derivative, E-M35, probably originated in eastern Africa.
…This inference is further supported by the presence of additional Hg E lineal diversification and by the highest frequency of E-P2* and E-M35* in the same region.
…The distribution of E-P2* appears limited to eastern African peoples.
…The E-M35* lineage shows its highest frequency (19.2%) in the Ethiopian Oromo but with a wider distribution range than E-P2*. Indeed, it is also found at high frequency (16.7%) in the Khoisan of South Africa (Underhill et al. 2000; Cruciani et al. 2002) (suggesting, once again, their ancient relationship with Ethiopians) and observed in southern Europe (present study).
…It is interesting that both E-P2* and E-M35* and their derivatives, E-M78 and E-M123, exhibit in Ethiopians the 12-repeat allele at the DYS392 microsatellite locus, an allele scarcely seen (Y-Chromosome STR Database), especially in other haplogroups and other populations (A.S.S.-B., unpublished data). In addition, the Ethiopian DYS392-12 allele is usually associated with the unusually short DYS19-11 allele, which is typical of this area.
…These findings are not easily explained. One possible scenario is that an ancient differentiation of the E-P2 haplogroup occurred in loco (East Africa). However, this also implies a low mutability of the associated microsatellite motif (DYS392-12/DYS19-11). Alternatively, the microsatellite motif may be due to HOMOPLASY.
The first scenario is more likely, since this unique microsatellite haplotype occurs in E-P2*, E-M35*, and E-M78 but is almost absent in all other haplogroups and populations.”
That is in Africa E3b is significantly different than that found outside of Africa, linking the so-called Sub-Saharan African to the Eastern African but not to Europe, Europeans, Eurasians, Jews, Arabs, or aliens!
No Hamitic-Semitic foundation. Eastern Africans are as ‘Black” or “Negroid” as any so-called Sub-Saharan African.
What is homoplasy?
http://www.sciencedaily.com/releases/2011/02/110224161512.htm
“For example, studying situations where a derived trait surfaces in two lineages that lack a recent common ancestor, or situations where an ancestral trait was lost but then reappeared many generations later, may help scientists identify the processes and mechanisms of evolution.”
Just because you say the Falasha are ‘mulattoes’ has no truth in it and clearly the genome diversity of Africans accounts for different morphologies/phenoypes that are independent of admixture and affected more by homoplasy!
CN
You wrote…
“My link showing the Jewish paternal haplotypes(Very likely, Aran influence to – since Jews and Arabs share 40% of the same genetic material) in Black Ethiopians is from 2010, more recent than ANY of your data. The data in your last response i svery mixed, even contradicting itself in many places – yes, I know, SUPPOSEDLY written by TOP NOTCH experts, who, incidentally, have changed their terms many times so it appears they are say something, but meaning something else – ESPECIALLY, haplotype V, M(35), or whatever other term they use.”
Of the 87 references you cited not one is a 2010 study. So why not provide that link?
By the way do you know how many subclades are in Haplogroup E?
http://www.ncbi.nlm.nih.gov/pubmed/8935325
Hum Biol. 1996 Jun;68(3):467-71.
Haplotype VIII of the Y chromosome is the ancestral haplotype in Jews.
Lucotte G, David F, Berriche S.
Source
International Institute of Anthropology, Paris, France.
Abstract
DNA samples from Ashkenazic and Sephardic Jews were studied with the Y-chromosome-specific DNA probes p49f and p49a to screen for restriction fragment length polymorphisms and haplotypes. Two haplotypes (VII and VIII) are the most widespread, representing about 50% of the total number of haplotypes in Jews. The major haplotype in Oriental Jews is haplotype VIII (85.1%); haplotype VIII is also the major haplotype in the Djerban Jews (77.5%) (Djerban Jews represent probably one of the oldest Jewish communities). Together these results confirm that HAPLOTYPE VIII is the ancestral haplotype in Jews.
NOT HAPLOTYPE V!!!!!!!!!!!!
CN…
Your quote:
“It is important to consider more complex models of population genesis, which allow for historically visible “groups” to be heterogeneous at origin, due to evolutionary (or social) processes, instead of interpreting heterogeneity as a necessary sign of admixture between distinct historically-known groups with different haplotypes or gene frequencies. Also models can be explored that postulate populations to be a blend of different historically known (or reported) ethno-ancestral groups, yet be genetically relatively “homogeneous,” as well as those that have a known (or reported) single ethnic origin, but yet are genetically “heterogeneous.”
Obviously, the time depth of “origins” and what this means must be carefully defined. Flexibility in model-building may help interpret situations that may be foreign to our current conceptions and paradigms. It is possible for a biologically-defined group to change cultural-linguistic [End Page 240] identities due to adoption/language shift, and for a cultural-linguistically defined/maintained community to change biologically because it diachronically and bioculturally assimilates numerous individuals who were GENETICALLY and/or MORPPOLOGICALLY DIFFERENT.”
:) http://www.ncbi.nlm.nih.gov/pubmed/10592688
Performing your original search, Falasha Jews, in PubMed will retrieve 2 records.
Hum Biol. 1999 Dec;71(6):989-93.
Origins of Falasha Jews studied by haplotypes of the Y chromosome.
Lucotte G, Smets P.
Source
International Institute of Anthropology, Paris, France.
Abstract
DNA samples from Falasha Jews and Ethiopians were studied with the Y-chromosome-specific DNA probe p49a to screen for TaqI restriction polymorphisms and haplotypes.
…Two haplotypes (V and XI) are the most widespread in Falashas and Ethiopians, representing about 70% of the total number of haplotypes in Ethiopia.
…Because the Jewish haplotypes VII and VIII are not represented in the Falasha population, we conclude that the Falasha people descended from ancient inhabitants of Ethiopia who converted to Judaism.”
It cannot get any clearer these are not of the same genetic strain as the so-called ‘White-Semitic-Jewish/Arab’ haplotypes.
They do not become ‘mulattoes’ because you say they are admixted when they are not.
But…you can say anything…
The Skeptioc
QUOTEING YOU, “I never said that the Ancient Egyptians were not Nubians – which seems to be your point.” Correct – keep INSISTING/INDICATING they were Black NUBIANS – I keep proving they were NOT.
I NEVER said this – “I never said that the “Black Jews” were not phenotyically ‘African’. In fact, are Afrasian, Afro-Asiatic and previously stated! You seem to think that this admixture ‘MAKES” them WHITE! Wrong!” – I said they were MIXED as in Mulattoes.The Falsha – as shown in my link from their paternal lineage – Jewish Semitic White ( VERY likely, Arab Semitic White to).
The most common variants found in different studies of Egypt collectively are, in descending frequency, V, XI, IV, VII, VIII, XV, and XII (Table 2A). The first three of these are of greatest interest due to their frequencies. Haplotype V, sometimes called “Arabic” (Lucotte and Mercier 2003a) declines from lower Egypt (north) at 51.9%, to upper Egypt (24.2%), and to lower Nubia (south) at 17.4%. Haplotypes VII, VIII, XV, and XII also decline (Table 1). In contrast, haplotypes XI and IV, called “southern,” with IV being labeled “sub-Saharan,” have their lowest frequencies in lower Egypt (XI-11.7%; IV-1.2%), but increase in upper Egypt (XI-28.8%; IV-27.3%); and lower Nubia (XI-30.4%; IV-39.1%); there is no statistically significant difference between the latter [End Page 223] two regions (Lucotte and Mercier 2003a). Haplotypes VII and VIII are most prevalent in the Near East, and XII and XV in Europe.
Summary of the most common p49a,f TaqI Y-chromosome haplotypes in Egyptian regions (Lucotte and Mercier 2003a).
Click for larger view Table 1
Summary of the most common p49a,f TaqI Y-chromosome haplotypes in Egyptian regions (Lucotte and Mercier 2003a).
Frequencies of p49a,f TaqI Y-chromosome haplotypes in selected African and Near Eastern countries (published data).
Click for larger view Table 2A
Frequencies of p49a,f TaqI Y-chromosome haplotypes in selected African and Near Eastern countries (published data).
It is important to address the appellation of “Arabic” for haplotype V, due to names being interpreted as indicators of origins, and the inconsistencies found in the literature. This variant is found in very high frequencies [End Page 224] in supra-Saharan countries and Mauretania (collective average 55.0%), and in Ethiopia (average 45.8%) (Table 2A). In specific groups its highest prevalence is in samples from Moroccan Amazigh (Berbers) (68.9%) and Ethiopian Falasha (60.5%). Its frequency is considerably less in the Near East, and decreases from west (Lebanon, 16.7%) to east (Iraq, 7.2%) (Table 2A). The label “Arabic” for V is therefore misleading because it suggests a Near Eastern origin. In fact this variant has been called “African” (Lucotte et al. 1993:839, Lucotte et al. 1996:469), and “Berberian” (Lucotte et al. 2001:887).
TaqI p49a, f Y-chromosome haplotype frequencies in more restricted populations from the Horn, supra-Saharan Africa, and of Near East origin (published data).
diversification and migration are conceptualized in Saharan and supra-Saharan Africa. It is important to state that most archeologists do not interpret the Capsian as being of Near Eastern origin.
Haplotype IV, designating the M2/PN1 subclade, as noted, is found in high frequency in west, central, and sub-equatorial Africa in speakers of Niger-Congo—which may have a special relationship with Nilosaharan—spoken by Nubians; together they might form a superphylum called Kongo-Saharan or Niger-Saharan (see Gregersen 1972, Blench 1995), but this is not fully supported. The spatial distribution of p49a,f TaqI haplotypes in the geographically-widespread speakers of Nilosaharan languages [End Page 228] has not been fully characterized, but the notable presence of haplotype IV in Nubians speaking the Eastern Sudanic branch is interesting in that this subgroup is in the Sahelian branch of speakers, whose ancestors may have participated in the domestication of cattle in the eastern Sahara (Ehret 2000, Wendorf and Schild 2001). Sometimes haplotype IV (and the M2 lineage) is seen as being associated with the “Bantu expansion” (~2000-3000 bp), but this does not mean that it is not much older, since expansion and origin times cannot be conflated. Haplotype IV has substantial frequencies in upper Egypt and Nubia, greater than VII and VIII, and even V. Bantu languages were never spoken in these regions or Senegal, where M2 is greater than 90 percent in some studies.
Haplotype XI has its highest frequencies in the Horn and the Nile valley, but has been called “Oriental” (see Lucotte for this appellation 1996:469), which is also misleading. The high frequency in both Nilosaharan and Afroasiatic speakers in northeast Africa is striking. This haplotype has arisen independently several times as indicated by its affiliation with lineages defined by different biallelic markers (see e.g., al-Zahery 2003, O. Semino, personal communication). The notable frequency in Askenazic Jews is likely primarily of European, not African, origin (the EU19 lineage, see Passarino et al. 2000), or in haplogroup R (see al-Zahery 2003). In northwestern Africa (Tunisia) it is likely to be of both African and European origin due to the region’s populations’ various historical interactions with Europe.
The widespread distribution of the PN2 clade in the major language phyla of Africa, its existence in the Levantine-Iraq region and even in the Aegean, and its likely post-glacial maximum date are significant and show how numerous bioculturally diverse peoples can be connected, even at relatively shallow time depths. This should give pause to those who have trouble escaping racial thinking. The diversification and early expansion of PN2 bearing populations likely started in the northeast quadrant of Africa (defined by bisecting the continent along its north-south axis and at the equator). This region is postulated to be the ancestral home of two of the three major language phyla of supra-equatorial Africa: Nilosaharan and Afroasiatic (Blench 1993, Ehret 1984, personal communication).
It is significant that bearers of the PN2 mutation are geographically widespread and diverse in external morphology and language family affiliation. There is also biological diversity even within the speakers of language families (in their “homelands”) that could be seen by some as problematic. The range of external morphologies in the continental African speakers of Afroasiatic cannot be viewed as problematic from an evolutionary [End Page 229] (versus racio-typological) perspective, and indicates the richness and complexity of indigenous African biocultural microevolution and its diversity (Hiernaux 1974, Keita and Kittles 1997, Kittles and Keita 1999). Conceptual racio-typological approaches that only interpret variation in terms of the interaction of primordial pre-existing distinct biocultural units will not easily explain phenomena like the PN2 distribution.
Accepting even the lower putative age of the mutation (Hammer and Zegura 2002 vs. Bosch et al. 2001), and language phyla (Ehret 1984), it can be suggested that PN2 and descendants perhaps arose in a population that antedates these language groupings, and which later heavily contributed to, or became the biopopulation base of, the nascent speech communities. Alternatively, it could mean that there was extensive interaction between the speakers of the ancestral linguistic families, postulating that the descendant mutations arose in these, with a subsequent different distribution in populations of various speech families. Haplotypes V and XI are somewhat ubiquitous in African language families (see Poloni 1997). In either case it is likely that a very successful subsistence strategy in the northeast quadrant of Africa made this possible (see e.g., Connor and Marks 1986, Wetterstrom 1993).
As noted, VII and VIII are the major indigenous Near Eastern haplotypes, and found to predominate in extant core descendant communities: Near Eastern Arabic speakers and Jews. In comparison to those of V their frequencies are small in supra-Saharan Africa (Tables 2A, 2B). Again employing the Falasha and northern Africa cases as a models, and the genetic evidence, it can be postulated that selected M35 carriers, speakers (from Africa) of a stage of ancestral Semitic (pre-proto-Semitic) entered the Near East, where indigenous peoples adopted it, and via ongoing language shift and population growth eventually became numerically greater than the original speakers of the ancestor.
As noted with reservation, the archeological “signal” for such movement might be the presence of the Mushabi industry in the Levant that has Nile Valley affinities (Bar-Yosef 1987, Midant-Reynes 2000). The large number of Mushabi sites suggests a major migration (see comments in Bar-Yosef 1987). However, this can only be tentatively suggested because there may be little concordance between language family and the distribution of archeological artifacts. Also the Mushabi may be indigenous to the Levant. The point is that an African proto-language grouping was adopted by indigenous Near Eastern peoples, based on linguistics and genetics. Eventually attestable Semitic emerged; reconstruction of this Common Semitic indicates that its speakers were food producers and not hunters and gatherers, as were the speakers of undifferentiated Afroasiatic [End Page 230] (see Diakonoff 1981, and revision 1998, Ehret 1984, 1995, personal communication).
Later there is some movement into Africa after the domestication of plants and ovacaprines, which happened in the Near East nearly 2000 years before it occurred in Egypt (Hassan 1988, Wetterstrom 1993). Early neolithic levels in northern Egypt contain the Levantine domesticates, and show some influence in material culture as well (Kobusiewicz 1992). Ovacaprines appear in the western desert before the Nile valley proper (Wendorf and Schild 2001). However, it is significant that the ancient Egyptian words for the major Near Eastern domesticates—sheep, goat, barley, and wheat—are not loans from either Semitic, Sumerian, or Indo-European. This argues against a mass settler colonization (at replacement levels) of the Nile valley from the Near East at this time. This is in contrast with some words for domesticates in some early Semitic languages, which are likely Sumerian loan words (Diakonoff 1981).
This evidence indicates that the northern Nile valley peoples apparently incorporated the Near Eastern domesticates into a Nilotic foraging subsistence tradition on their own terms (Wetterstrom 1993). There was apparently no “neolithic revolution” brought by settler colonization, but a gradual process of neolithicization (Midant-Reynes 2000). While some Neolithic movement took place, there is the problem of sifting the results of this from later migration. (Also some of those emigrating may have been carrying haplotype V, descendents of earlier migrants from the Nile valley, given the postulated “Mesolithic” time of the M35 lineage emigration). It is more probable that the current VII and VIII frequencies, greatest in northern Egypt, reflect in the main (but not solely) movements during the Islamic period (Nebel et al. 2002), when some deliberate settlement of Arab tribes was done in Africa, and the effects of polygamy. There must also have been some impact of Near Easterners who settled in the delta at various times in ancient Egypt (Gardiner 1961). More recent movements, in the last two centuries, must not be forgotten in this assessment.
The mode and patterns of migration discussed above would account for the opposing east-west clines of V versus VII+VIII in southwest Asia, and the higher frequency of V nearer Africa (Egypt). The Greco-Roman incursions (Gardiner 1961) are the earliest text-supported migrations that may account for XII and XV in Egypt. There is little evidence for earlier movements, but these likely did occur to Egypt’s Mediterranean coast. Both of these haplotypes have high frequencies in Europe (Perischetti et al. 1992, Lucotte and Loriat 1999), and are found on different biallelic lineages than the most frequent haplotypes found in Egypt (cf. Hammer and Zegura 2002, Underhill et al 2001, al-Zahery et al 2003). [End Page 231]
The very noteworthy frequency of XII in Tunisia might reasonably in part be attributed to the settlement of numbers of Roman soldiers and administrators and their families after the defeat of Carthage, perhaps increased by some form of sexual or social selection. There was also likely “Copper-Age” migration from Sardinia (Camps 1982), and ongoing contact with nearby islands in the Mediterranean. Somewhat surprising for Tunisia is the relative paucity of VII+VIII given the Phoenician settler colonies, and its later role in the Islamic period. This is likely due to sampling since other studies suggest a larger Near Eastern impact (Hammer, personal communication).
This is a reminder that genes, languages, and nationalities are not intrinsically linked, and that numerous samples would be helpful in getting an accurate assessment. The well-known Greek colonies in urban Cyrenaica (in modern Libya) also must not be forgotten in this regard, as well as the reflux of European converts to Islam back into Africa, after the expulsion of Jews and Muslims from Europe in the fifteenth century. This last event might account for the frequency of haplotype XV in Morocco.
III
The data for Egypt, north to south, are rendered more interesting in light of the distributions in adjacent regions. The high prevalence of V in Ethiopia, south of Egypt, would alone seem to indicate that movements associated with Dynasty XII and XVIII Egyptian military colonizations are not sufficient explanations for frequencies in lower Nubia and upper Egypt, statistically the same. The decreasing cline does not continue. Ethiopian (and Falasha) frequencies are higher than in upper Egypt. This observation is not the case for haplotypes VII, VIII, XII, and XV, although, ironically, haplotype VIII has a notable presence in a sample of non-Falasha Ethiopians from north of Addis Ababa (Lucotte and Smets 1999). TRANSLATION (mine) – Moving from Lower Egypt(Northern Egypt) to Upper Egypt (Southern Egypt) – the population goes fro, White(Semitic), to Black(Nubian)
Leaving aside the smaller frequencies of the “European” haplotypes, and the likely migrations associated with them (see Lucotte and Mercier 2003a), what other interactions may help explain the patterns of the distributions of V, XI, IV, VII, and VIII in Africa and southwest Asia (the Near East)? What were their pre-Middle Kingdom frequencies in the Egyptian Nile valley, and what events may have helped shape them? We hypothesize that early holocene settlement and population interactions, not later military incursions, are the major mechanisms that accounts for the haplotype patterns, and that prevalence locates their most parsimonious [End Page 232] geographical sources, assuming a minimal number of unusual founder, expansion, and extinction events.
It is possible that the spread of the haplotypes bears some relationship to the spread of language families. Recall that the languages spoken in the Nile valley, Horn, and supra-Saharan Africa west of Egypt, as well as the central and southern Sahara, belong primarily to the Afroasiatic and Nilo-Saharan phyla (or families) (Greenberg 1966, Ehret 1984, Ruhlen 1987). Nubian in the Nile valley is Nilo-Saharan. Ancestral (proto-)Afroasiatic may date from 15,000 to 13,000 BCE (Ehret 1984), or more. Its differentiation through space and time and movement occurred primarily in Africa, producing at least six families: Omotic, Cushitic, Chadic, ancient Egyptian, Berber, and Semitic. In a phylogenetic model these last four are concluded to be the “younger” members of the family, but the nature of the process of linguistic differentiation might make certain dating difficult.
Hypotheses that bring Afroasiatic from Asia or Europe with agriculture are not parsimonious (Ehret, personal communication). The Nostratic hypothesis that proffers this view has largely been modified and abandoned; most Nostraticists now see Afroasiatic as a sister of Nostratic and not a daughter (Ruhlen 1991). The common parent to these would reach back into a time not generally believed to be validly accessible to standard linguistic methods (Nichols 1997), although there is dissent on this point.
The distribution and high prevalence of haplotype V (and less so of XI, Nile valley primarily), and Afroasiatic speakers in Africa correspond with the geography of the Horn-supra-Saharan arc. This is suggestive. The spread of the language phylum and genes may illustrate a case of kin-structured migration (Fix 1999), with founder-effect in some instances (e.g., high frequency of V in Moroccan Berbers). In the southern Nile valley V (and XI) might have been established with early Afroasiatic speakers, whose reconstructed vocabulary on available evidence suggests that they were hunters and intensive plant users, not food producers (see Ehret 1988, 2000, for a discussion of cultural reconstruction from language, and Ehret 1984).
This subsistence pattern characterizes a late paleolithic site from Wadi Kubanniya in southern Egypt (Wetterstrom 1993), and subsequent epipaleolithic sites. Early Afroasiatic speakers, along with those of Nilosaharan, were likely drawn into the Sahara, which was less arid in the late pleistocene in the early holocene after the last glacial maximum. Over time, as Afroasiatic differentiated and populations migrated, founder effect with kin-structured migration may have led to the basic distribution of V seen in the Horn and northern Africa today. Haplotype V has a [End Page 233] much lower frequency among core Semitic-speaking descendant communities in the Near East (i.e., Arabs and Jews).
It should be reiterated that using the same logic as applied to assess the Falasha, and the Arabic speakers of supra-Saharan Africa, it can be postulated that the ancestor of undifferentiated Semitic was adopted in the Near East by peoples having a prevalence of haplotypes VII and VIII. The levels and cline of V in the region are consistent with this hypothesis. Haplotype V in northern Egypt may also have had recurrent sources: in addition to a neolithic return of some having haplotype V, the Libyan kings of dynasties XXII-XXIV (~950-750 BC), based in the delta, might also have settled their countrymen. These would have been Amazigh (Berber)-speaking populations probably with a predominant frequency of haplotype V. It is difficult to judge the impact of these.
Archeological data, or the absence of it, have been interpreted as suggesting a population hiatus in the settlement of the Nile Valley between the epipaleolithic and the neolithic/predynastic, but this apparent lack could be due to material now being covered over by the Nile (see Connor and Marks 1986, Midant-Reynes 2000, for a discussion). Analagous to events in the Atacama Desert in Chile (Nuñez et al 2002), a moister more inhabitable eastern Sahara gained more human population in the late pleistocene-early holocene (Wendorf and Schild 1980, Hassan 1988, Wendorf and Schild 2001). If the hiatus was real then perhaps many Nile populations became Saharan.
Later, stimulated by mid-holocene droughts, migration from the Sahara contributed population to the Nile valley (Hassan 1988, Kobusiewicz 1992, Wendorf and Schild 1980, 2001); the predynastic of upper Egypt and later neolithic in lower Egypt show clear Saharan affinities. A striking increase of pastoralists’ hearths are found in the Nile valley dating to between 5000-4000 BCE (Hassan 1988). Saharan Nilosaharan-speakers may have been the initial domesticators of African cattle found in the Sahara (see Ehret 2000, Wendorf et. al. 1987). Hence there was a Saharan “neolithic” with evidence for domesticated cattle before they appear in the Nile valley (Wendorf et al. 2001). If modern data can be used, there is no reason to think that the peoples drawn into the Sahara in the earlier periods were likely to have been biologically or linguistically uniform.
Conceptually, modeling the early to mid-holocene eastern Sahara, including the Nile valley, as being the locale of a metapopulation in a deteriorating habitat, and undergoing reduction from dispersal might help explain the current Nile valley diversity (see Gyllenberg and Hanski 1997, Gandon and Michalakis 1999, Hanski and Ovaskainen 2000, Duncan et [End Page 234] al. 2001, Poethke and Hovestadt 2002, Nuñez et al. 2002). A dynamic diachronic interaction consisting of the fusion, fissioning, and perhaps “extinction” of populations, with a decrease in overall numbers as the environment eroded, can easily be envisioned in the heterogeneous landscape of the eastern Saharan expanse, with its oases and wadis, that formed a reticulated pattern of habitats. This fragile and changing region with the Nile valley in the early to mid-holocene can be further envisioned as holding a population whose subdivisions maintained some distinctiveness, but did exchange genes. Groups would have been distributed in settlements based on resources, but likely had contacts based on artefact variation (Wendorf and Schild 2001). Similar pottery can be found over extensive areas. Transhumance between the Nile valley and the Sahara would have provided east-west contact, even before the later migration that largely emptied parts of the eastern Sahara.
Early speakers of Nilosaharan and Afroasiatic apparently interacted based on the evidence of loan words (Ehret, personal communication). Nilosaharan’s current range is roughly congruent with the so-called Saharo-Sudanese or Aqualithic culture associated with the less arid period (Wendorf and Schild 1980), and therefore cannot be seen as intrusive. Its speakers are found from the Nile to the Niger rivers in the Sahara and Sahel, and south into Kenya. The eastern Sahara was likely a micro-evolutionary processor and pump of populations, who may have developed various specific sociocultural (and linguistic) identities, but were genealogically “mixed” in terms of origins.
These identities may have further crystallized on the Nile, or fused with those of resident populations that were already differentiated. The genetic profile of the Nile Valley via the fusion of the Saharans and the indigenous peoples were likely established in the main long before the Middle Kingdom. Post-neolithic/predynastic population growth, as based on extrapolations from settlement patterns (Butzer 1976) would have led to relative genetic stability. The population of Egypt at the end of the predynastic is estimated to have been greater than 800,000, but was not evenly distributed along the valley corridor, being most concentrated in locales of important settlements (Butzer 1976). Nubia, as noted, was less densely populated.
Interactions between Nubia and Egypt (and the Sahara as well) occurred in the period between 4000 and 3000 BCE (the predynastic). There is evidence for sharing of some cultural traits between Sudan and Egypt in the neolithic (Kroeper 1996). Some items of “material” culture were also shared in the phase called Naqada I between the Nubian A-Group and upper Egypt (~3900-3650 BCE). There is good evidence for a [End Page 235] zone of cultural overlap versus an absolute boundary (Wilkinson 1999 after Hoffman 1982, and citing evidence from Needler 1984 and Adams 1996). Hoffman (1982) noted cattle burials in Hierakonpolis, the most important of predynastic upper Egyptian cities in the later predynastic. This custom might reflect Nubian cultural impact, a common cultural background, or the presence of Nubians.
Whatever the case, there was some cultural and economic bases for all levels of social intercourse, as well as geographical proximity. There was some shared iconography in the kingdoms that emerged in Nubia and upper Egypt around 3300 BCE (Williams 1986). Although disputed, there is evidence that Nubia may have even militarily engaged upper Egypt before Dynasty I, and contributed leadership in the unification of Egypt (Williams 1986). The point of reviewing these data is to illustrate that the evidence suggests a basis for social interaction, and gene exchange.
There is a caveat for lower Egypt. If neolithic/predynastic northern Egyptian populations were characterized at one time by higher frequencies of VII and VIII (from Near Eastern migration), then immigration from Saharan sources could have brought more V and XI in the later northern neolithic. It should further be noted that the ancient Egyptians interpreted their unifying king, Narmer (either the last of Dynasty 0, or the first of Dynasty I), as having been upper Egyptian and moving from south to north with victorious armies (Gardiner 1961, Wilkinson 1999). However, this may only be the heraldic “fixation” of an achieved political and cultural status quo (Hassan 1988), with little or no actual troup/population movements. Nevertheless, it is upper Egyptian (predynastic) culture that comes to dominate the country and emerges as the basis of dynastic civilization. Northern graves over the latter part of the predynastic do become like those in the south (see Bard 1994); some emigration to the north may have occurred—of people as well as ideas.
Interestingly, there is evidence from skeletal biology that upper Egypt in large towns at least, was possibly becoming more diverse over time due to immigration from northerners, as the sociocultural unity proceeded during the predynastic, at least in some major centers (Keita 1992, 1996). This could indicate that the south had been impacted by northerners with haplotypes V, VII, and VIII, thus altering southern populations with higher than now observed levels of IV and XI, if the craniometric data indicate a general phenomenon, which is not likely. The recoverable graves associated with major towns are not likely reflective of the entire population. It is important to remember that population growth in Egypt was ongoing, and any hypothesis must be tempered with this consideration. [End Page 236]
Dynasty I brought the political conquest (and cultural extirpation?) of the A-Group Nubian kingdom Ta Seti by (ca. 3000 BC) Egyptian kings (Wilkinson 1999). Lower Nubia seems to have become largely “depopulated,” based on archeological evidence, but this more likely means that Nubians were partially bioculturally assimilated into southern Egypt. Lower Nubia had a much smaller population than Egypt, which is important to consider in writing of the historical biology of the population. It is important to note that Ta Seti (or Ta Sti, Ta Sety) was the name of the southernmost nome (district) of upper Egypt recorded in later times (Gardiner 1961), which perhaps indicates that the older Nubia was not forgotten/obliterated to historical memory.
Depending on how “Nubia” is conceptualized, the early kingdom seems to have more or less became absorbed politically into Egypt. Egypt continued activities in Nubia in later Dynasty I (Wilkinson 1999, Emery 1961). A different reading of the documents interpreted as indicating the defeat of Nubia by Dynasty I kings is that these rulers were defending Nubian allies who had assisted them in consolidating Egypt from attacks by other Nubians (see Trigger 1976). Over the dynastic period Nubians were continuously brought into Egyptian armies as mercenaries—sometimes even to fight other Nubians (Trigger 1976). There was steady Nubian contact, especially in upper Egypt. Nubians were allegedly carried off into Egypt in great numbers during the Old Kingdom (Dynasties III to VI). (Emery 1961, Wilkinson 1999). In the First Intermediate Period Nubian mercenaries assimilated into the Upper Egyptian population (Fischer 1961).
In later times it was also kings or leaders from the south, with southern armies and sometimes Nubian mercenaries, who restored unity to Egypt; this was the case for the Dynasty XI, whose rulers made possible the Middle Kingdom, and whose pharaohs subsequently also raided Nubia, establishing forts there and an apparently small presence. Middle Kingdom forts did not hold large populations (Trigger 1976). It also seems likely that C-Group Nubian population and culture “disappears” because of biocultural assimilation into Upper Egyptian society in the Second Intermediate Period (Hafsaas, 2004). This is another possible source of variation assuming that they were different in the first place.
In the tradition of southern Egyptian leaders, the later Nubian kings (Dynasty XXV) who conquered Egypt saw themselves as restorers and revivalists in some sense, and not apparently as foreigners; this would have likely influenced their behavior toward ordinary Egyptians. Evidence for this is found in the Victory Stela of Pi(ankhy), founder of Dynasty XXV; the text does not suggest an attitude seeking settler colonization or [End Page 237] territory (see translations by Lichtheim 1980, Goedicke 1998). It is worth noting that during the Islamic period that Christian Nubians sometimes controlled, or had great influence in, upper Egypt (Shinnie and Shinnie 1965).
The New Kingdom, which was made possible by Dynasty XVII southern upper Egyptians who expelled the Hyksos, later conquered and effectively colonized lower and upper Nubia to the fourth cataract. Lower Nubia was not the threat, but rather the kingdom of Kush, whose rulers had allied themselves with the Asiatic Hyksos between the Middle and New Kingdoms. This colonization lasted 500 years, to the end of the New Kingdom. There was an Egyptianization of Nubian elites that later extended to the masses, and Egyptians were even settled deep in upper Nubia. Prisoners and enslaved locals were sometimes sent to Egypt and settled there (Trigger 1976), but it is difficult to quantify the number of translocated persons. No doubt some assimilated individuals also went to Egypt.
After the New Kingdom, Egyptians either returned home or simply fused with the local population. In contrast to Egyptian New Kingdom colonization, the Nubian control of Egypt was less than 100 years in duration, and there is no record of a program of settler colonization. Given the Egyptian versus Nubian actions it is striking how small the percentage of V in Nubia is, versus IV and XI in upper Egypt (Table I), if these military events alone are viewed as being responsible for extant regional genetic profiles, and if these variants are treated as being ethnically specific.
Taking a long and synthetic view, one compelling scenario is as follows: after the early late pleistocene/holocene establishment of Afroasiatic-speaking populations in the Nile valley and Sahara, who can be inferred to have been predominantly, but not only V (and XI), and of Nilosaharan folk in Nubia, Sudan, and Sahara (mainly XI and IV?), mid-holocene climatic-driven migrations led to a major settlement of the valley in upper Egypt and Nubia, but less so in lower Egypt, by diverse Saharans having haplotypes IV, XI, and V in proportions that would significantly influence the Nile valley-dwelling populations.
These mid-Holocene Saharans are postulated to have been part of a process that led to a diverse but connected metapopulation. These peoples fused with the indigenous valley peoples, as did Near Easterners with VII and VIII, but perhaps also some V. With population growth the genetic profiles would became stabilized. Nubian and upper Egyptian proximity and on some level, shared culture, Nubia’s possible participation in Egyptian state-building, and later partial political absorption in Dynasty [End Page 238] I, would have reinforced biological overlap (and been further “stabilized” by ongoing population growth).
In this model much later migrations would have not created the genetic profile, only helped to maintain it. Although Nubia was occupied for some 500 years during the New Kingdom, there apparently was no genocidal settler colonization. However, there is evidence for the Egyptianization of Nubians and other enslaved southerners (Nubians proper and others) being taken to Egypt, but it is hard to imagine that the assimilation of these individuals would have greatly affected gene frequencies, all other things being equal. The relatively brief non-colonizing control of Egypt by Nubians would not have had the effect of a half millennium of occupation unless there was some specific policy of assimilation. These interactions, in the view advocated here, would have reinforced a basic genetic pattern long present in southern Egypt.
Considering the possible explanations for the Y variation, the clinal patterns observed for mtDNA variants (Krings et al. 1999) become subjects of interest. This DNA is usually only inherited maternally. The mtDNA variants’ distributions have been been used to interpret the Nile valley as a zone of intergradation, created by the admixture populations of distinct northern and southern origin having different haplotypes. Movement up and down the Nile corridor is the mechanism postulated to have produced the pattern (from the Mediterranean to the southern Sudan). The three military invasions have also been invoked to explain the mtDNA patterns (Krings et al 1999:1173). This is a less tenable explanation for these variants, since women were not soldiers in ancient Egypt and Nubia, and wives of soldiers would not likely have contributed to the gene pools of the conquered. The translocation of a lot of the population of the victorious parties is not attested.
However, the coalescence times for the slowly evolving northern and southernmost haplotypes by region should be considered (see Krings et. al. 1999). These would seem to place the ancestor in the epoch of the less arid Sahara, in the early to pre-mid-holocene, when it was more populated or shortly after, when droughts were influential in causing emigration. Hence it can be argued that the scenario presented for the Y chromosome variation—of Saharan interactions and migrations into the valley—and later events would also have some power in explaining the distributions of the mtDNA variants, at least in part. Differential bidirectional north-south migration by itself would not likely be the only explanation for the findings. One needs also to consider under what social circumstances would delta Egyptian women come to be in the southern Sudan, unless only the village-to-village transfer of DNA is postulated. [End Page 239]
The more recent upheavals in the Sudan may also have altered patterns. The social context/circumstances of gene flow must always be considered, and ideally understood. The historical linguistic data reported earlier would apply in the case of maternal lineages as well. It can also be argued that it is not likely that the “northern” genetic profile is simply due to “Eurasians” having colonized supra-Saharan regions from external African sources. It might be likely that the greater percentage of haplotypes called “Eurasian” are predominantly, although not solely, of indigenous African origin. As a term “Eurasian” is likely misleading, since it suggests a single locale of geographical origins. This is because it can be postulated that differentiation of the L3* haplogroup began before the emigration out of Africa, and that there would be indigenous supra-Saharan/Saharan or Horn-supra-Saharan haplotypes. More work and careful analysis of mtDNA and the archeological data and likely probabilities is needed. Early hunting and gathering paleolithic populations can be modeled as having roamed between northern Africa and Eurasia, leaving an asymmetrical distribution of various derivative variants over a wide region, giving the appearance of Eurasian incursion.
It is of some interest that the patterns observed in the Nile valley across ethno-national boundaries for both types of lineage DNA do not apparently conform to those found in idealized strictly patrilineal/patriarchal societies that admit diverse women to their ranks as mates, but exclude foreign males (Salem et al. 1996, al-Zahery 2003, Richards et al. 2003). The diversity in male and female lineages by regions is striking. This also justifies a more complex model of interpretation for the observed genetic variation beyond one that only considers linear migration in the Nile corridor, and exchange between formerly “pure” ethnopopulations.
It is important to consider more complex models of population genesis, which allow for historically visible “groups” to be heterogeneous at origin, due to evolutionary (or social) processes, instead of interpreting heterogeneity as a necessary sign of admixture between distinct historically-known groups with different haplotypes or gene frequencies. Also models can be explored that postulate populations to be a blend of different historically known (or reported) ethno-ancestral groups, yet be genetically relatively “homogeneous,” as well as those that have a known (or reported) single ethnic origin, but yet are genetically “heterogeneous.”
Obviously, the time depth of “origins” and what this means must be carefully defined. Flexibility in model-building may help interpret situations that may be foreign to our current conceptions and paradigms. It is possible for a biologically-defined group to change cultural-linguistic [End Page 240] iidentities due to adoption/language shift, and for a cultural-linguistically defined/maintained community to change biologically because it diachronically and bioculturally assimilates numerous individuals who were genetically and/or morphologically different.
In summary, late pleistocene, early and mid-holocene, and Dynasty I population movements that can be related to language family dispersals, Saharan aridity, droughts and Nile Valley settlement, mating patterns, social interactions other than warfare, as well as the effects of state-level conflicts should be integrated into discussions of Nile valley population histories.
This is generally applicable. Movements from the west and east to the Nile Valley, and north and south within the Nile corridor played a role in its population history. It is hypothesized that the events of the early settling of the Nile valley and interactions through Dynasty I and the Old Kingdom, and ongoing population growth, likely had as much of a role in generating the current Nile Valley pattern for the p49a,f TaqI Y haplotypes, as did events occurring in the Middle Kingdom and later. In this view these latter events, while contributory, were not the primary determinants of the distributions now observed. Future research, using computer simulation, might enable choosing the best model to explain the observed patterns of variation.
Bibliography
Adams, B (1996). “Elite graves at Hierakonpolis” in J. Spencer, ed. Aspects of Early Egypt. London. 1-15
Al-Zahery, N. et al (2003). “Y chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations.” Mol Phyl Evol 28:458-72
Angel, L. (1972). “Biological relationships of Egyptian and eastern Mediterranean populations during predynastic times.” Journal of Human Evolution 1:307-13
—. (1973) “Early Neolithic people of Nea Nikomedeia” in I. Schwidetsky I, ed. Fundamenta. Monographien zur Urgeschichte. Series B. Die Anfänge Neolithikums vom Orient bis Nordeuropa 3, pt. VIII. Cologne. 103-12
—. and J. Kelley (1986). “Description and comparison of the skeleton.” in F. Wendorf and R. Schild, eds. The Wadi Kubanniya Skeleton. Dallas. 53-70
Bard K. (1994). From Farmers to Pharaohs. Mortuary Evidence for the Rise of Complex Society. Edinburgh [End Page 241]
Bar-Yosef, O. (1987). “Pleistocene connexions between Africa and southwest Asia.” African Archaeological Review 5:29-30
Bender, L, (1975), Omotic: A New Afro-Asiatic Language Family. Carbondale
Blench, R. (1993). “Recent developments in African language classifications and their implications for prehistory” in T. Shaw et al., eds. The Archaeology of Africa: Food, Metal and Towns. New York, 126-37
—. (1995). “Is Niger-Congo simply a branch of Nilo-Saharan?” in R. Nicolai and F. Rottland F, eds., Proceedings of the Fifth Nilosaharan Linguistics Colloquium, Nice, 1992. Köln, 68-118
Bosch, E. et al. (2001). “High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian peninsula.” American Journal of Human Genetics 68:1019-29
Butzer, K.W. (1976). Early Hydraulic Civilization in Egypt. Chicago
Camps, G. (1982). “Beginnings of pastoralism and cultivation in northwest Africa and the Sahara: origins of the Berbers” in Cambridge History of Africa, I. Cambridge
Close, A. (1980-81). “The Iberomaurusian sequence at Tamar Hat.” Libyca 28/29: 69-104
Connor, D.R. and A. Marks (1986). “The terminal pleistocene on the Nile: the final Nilotic adjustment” in L.G. Straus, ed. The End of the Paleolithic in the Old World, 171-99
Cruciani, F. et al. (2002. “A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human y-chromosome haplotypes.” American Journal of Human Genetics 70:1197-1214
Duncan, S.R., C.J. Duncan, and S. Scott (2001). “Human population dynamics.” Annals of Human Biology 28:599-615
Diakonoff, I.M. (1981). “Earliest Semites in Asia.” Altorientalische Forschungen 8:23-74
—. (1998). “The Earliest Semitic society. Linguistic data.” Journal of Semitic Studies 4:209-19
Ehret, C. (1984). “Historical/linguistic evidence for early African food production” in J.D. Clark and S. Brandt, eds., From Hunters to Farmers. Berkeley, 26-36
—. (1988) “Language change and the correlates of language and ethnic shift.” Antiquity 62:564-74
—. (1993). “Nilosaharans and the Saharo-Sudanese Neolithic” in Shaw et al., Archaeology of Africa, 104-25 [End Page 242]
—. (1995). Reconstructing Proto-Afroasiatic (Proto-Afrasian): Vowels, Tone, Consonants and Vocabulary. Berkeley
—. (2000). “Language and history” in B. Heine and D. Nurse. African Languages: An Introduction. Cambridge, 272-97
Emery, W.B. (1961). Archaic Egypt. London
Fischer, H.G. (1961). “The Nubian Mercenaries of Gebelein During the First Intermediate Period.” Kush 9:44-84
Fix, A. (1999). Migration and Colonization in Human Microevolution. Cambridge
Fleming, H. (1974). “Omotic as an Afroasiatic family.” Studies in African Linguistics/ supplement 5:81-94
Gandon, S. and Y. Michalakis (1999). “Evolutionarily stable dispersal rate in a metapopulation with extinctions and kin competition.” Journal of Theoretical Biology 199:275-90
Gardiner, A. (1961). Egypt of the Pharaohs. Oxford
Goedicke, H. (1998). Pi(ankhy) In Egypt: A study of the Pi(ankhy) Stela. Baltimore
Greenberg, J.H. (1966). The Languages of Africa. Bloomington
—. (1973). “African languages” in E.P. Skinner, ed., Peoples and Cultures of Africa. Garden City, 34-58
Gregersen, E. (1972). “Kongo-Saharan.” Journal of African Linguistics 11:77-89
Gyllenberg, M. and I.I. (1997). “Habitat deterioration, habitat destruction, and metapopulation persistence in a heterogenous landscape.” Theoretical Population Biology 52:198-215
Hafsaas, H. (2004) “The C-Group People in the Crossfire. Lower Nubia during the Second Intermediate Period.” Paper presented at the SAFA annual meeting 2004. Bergen, Norway
Hammer, M. and S. Zegura (2002). “The human Y chromosome haplogroup tree: nomenclature and phylogeography of its major divisions.” Annual Review of Anthropology 31:303-21
Hanski, I. and O. Ovaskainen (2000) “The metapopulation capacity of a fragmented landscape.” Nature 404:755-58
Hassan, F. (1988). “The predynastic of Egypt.” Journal of World Prehistory 2:135-85
Hiernaux, J. (1974). The People of Africa. New York
Hoffman, M. (1982) The Predynastic of Hierakonpolis and Interim Report. Macomb.
Keita, S.O.Y. (1996). “Analysis of Naqada predynastic crania: a brief report.” in L. Kryaniak, K. Kroeper, and M. Kobusiewicz, eds. Interregional Contacts in the Later Prehistory of Northeastern Africa. Poznan, 205-14 [End Page 243]
—. (1992).” Further studies of crania from ancient northern Africa: an analysis of crania from First Dynasty Egyptian tombs, using multiple discriminant function.” American Journal of Physical Anthropology 87:245-54
— and R. Kittles (1997). “The persistence of racial thinking and the myth of racial divergence.” American Anthropologist 99:534-44
Kittles, R. and S.O.Y. Keita (1999). “Interpreting African genetic diversity.” African Archaeological Review 16:87-91
Kobusiewicz, M. (1992). “Neolithic and predynastic development in the Egyptian Nile Valley” in F. Klees and R. Kuper, eds. New Light on the Northeast African Past. Köln, 207-18
Krings, M. et al. (1999) “MtDNA analysis of Nile River Valley populations: a genetic corridor or a barrier to migration?” American Journal of Human Genetics 64:1166-76
Kroeper, K. (1996). “Minshat Abu Omar—burials with palettes” in J. Spencer, ed. Aspects of Early Egypt. London, 70-91
Lichtheim, M. (1980). Ancient Egyptian Literature: A Book of Readings, III, The Late Period. Berkeley
Lucotte, G., A. Aouizerate, and S. Berriche (2000). “Y-chromosome DNA haplotypes in North African populations.” Human Biology 72:473-80
—, F. David, and S. Berriche (1996). “Haplotype VIII of the Y chromosome is the ancestral haplotype in Jews.” Human Biology 68:467-71
—, N. Gerard, and G. Mercier (2001). “North African genes in Iberia studied by Y-chromosome DNA haplotype V.” Human Immunology 62:885-88
— and F. Loirat (1999). “Y-chromosome DNA haplotype 15 in Europe. Human Biology 71:431-37
— and G. Mercier (2003a). “Y-chromosome haplotypes in Egypt.” American Journal of Physical Anthropology 121:63-66
— and G. Mercier (2003b). “Y chromosome DNA haplotypes in Jews: comparisons with Lebanese and Palestinians.” Genetic Testing 7:67-71
— and P. Smets (1999). “Origins of Falasha Jews studied by haplotypes of Y chromosome.” Human Biology 71:989-93
—, P. Smets, and J. Ruffie (1993). “Y-chromosome-specific haplotype diversity in Ashkenazic and Sephardic Jews.” Human Biology 65:835-40
— et al. (1990). “The p49/ Taq I Y-specific polymorphisms in three grops of Indians.” Gene Geography 4:21-28
Manni, F. et al. 2002. “Y chromosome analysis in Egypt suggests a genetic regional continuity in northeastern Africa.” Human Biology 74:645-58 [End Page 244]
McBurney, C.B.M. (1960). The Stone Age of Northern Africa. London
Midant-Reynes, B. (2000). The Prehistory of Egypt from the First Egyptians to the First Pharaohs. Oxford
Munro-Hay, S. (1991). Aksum: An African Civilization of Late Antiquity. Edinburgh
Nebel, A. et al. (2002) Genetic evidence for the expansion of Arabian tribes into the southern Levant and North Africa. American Journal of Human Genetics 70:1594-96
Needler, W. (1984). Predynastic and Archaic Egypt in the Brooklyn Museum. Brooklyn
Ngo, K.Y. et al. (1986). “A DNA probe detecting multiple haplotypes of the human Y chromosome. American Journal of Human Genetics 38:407-18
Nichols, J. (1997). “Modeling ancient population structures and movement in linguistics.” Annual Review of Anthropology 26:359-84
Nuñez, L, M. Grosjean, and I. Cartajena (2002) “Human occupations and climate change in the Puna de Atacama, Chile.” Science 298:821-24
Passarino, G. et al. (2001). “The 49a,f haplotype 11 is a new marker of the EU19 lineage that traces migrations from northern regions of the Black Sea.” Human Immunology 62:922-32
Persichetti, F. et al. (1992). “Disequilibrium of multiple DNA markers on the human Y chromosome.” Annals of Human Genetics 56:303-310
Phillipson, D.W. (1985). African Archaeology. Cambridge
Poethke, H.J. and T. Hovestadt (2002). “Evolution of density- and patch-size-dependent dispersal rates.” Proceedings of the Royal Society London: B Biological Sciences 269: 637-45
Poloni, E.S. et al. (1997). “Human genetic affinities for Y-chromosome p49a,f TaqI haplotypes show strong correspondences with linguistics.” American Journal of Human Genetics 61:1015-1031
Richards, M. et al. (2003). “Extensive female-mediated gene flow from sub-Saharan Africa into near eastern Arab populations. American Journal of Human Genetics 72:1058-64
Rosser, Z.H. et al. (2000). “Y-chromosomal diversity in Europe is clinal and influenced primarily by geography than by language. American Journal of Human Genetics 67: 1526-43
Ruhlen, M. (1987). A guide to the World’s Languages. 1. Classification. Stanford
Salem, A.H. et al. (1996). “The genetics of traditional living: Y chromosomal and mitochondrial lineages in Sinai Peninsula.” American Journal of Human Genetics 59:741-74 [End Page 245]
Santichiara-Benerecetti, A.S. et al. (1993). “The common, Near Eastern origin of Ashkenazi and Sephardi Jews supported by Y-chromosome similarity.” Annals of Human Genetics 57:55-64
Shinnie, P.L. and M. (1965). “New light on medieval Nubia.” Journal of African History 6 275-94
Smith, P.E.L. (1982). “The Late Palaeolithic and Epi-Palaeolithic of northern Africa” in Cambridge History of Africa 1:342-409
Trigger, B. (1976). Nubia Under the Pharaohs. Boulder
Underhill P.A. et al. (2001). “The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Annals of Human Genetics 65:43-62
Wendorf, F., A.E. Close, and R. Schild (1987). “Early domestic cattle in the eastern Sahara” in Palaecology of Africa and the Surrounding Islands 18. Rotterdam, 441-48.
— and R. Schild (1980). Prehistory of the Eastern Sahara. New York
—. (2001). Holocene Settlement of the Egyptian Sahara, I. The Archaeology of Nabta Playa. New York
Wetterstrom, W. (1993). “Foraging and farming in Egypt: the transition from hunting and gathering to horticulture in the Nile Valley” in Shaw et al., Archaeology of Africa 165-226
Wilkinson, T.A.H. (1999) Early Dynastic Egypt. London
Williams, B.B. (1986). Excavations Between Abu Simbel and the Sudan Frontier. The A-Group Royal Cemetery at Qustul: Cemetery L. Chicago
Footnote
1. Discussions over a long period of time with G.A. Harrison provided useful input for the outline of this presentation, as did John Baines. P. Underhill provided useful comments that improved this manuscript. I wish to also thank F. Wendorf, F. Hassan, C. Ehret, A. Brooks, and R. Kittles, and the numerous participants at the Poznan Symposium on the Archaeology of Northeast Africa who shared data about the early Sahara, especially M. Kobusiewicz. Much more collaboration is planned with them. This piece is dedicated to John Baines, Professor of Egyptology at Oxford University, a supervisor and friend who provided much insight into ideas about, and the workings of, ancient Egypt. I would also recall the life and work of the late Larry Angel, another of my teachers, who was one pioneer in the synthesis of biological, linguistic, historical, and cultural data.
My link showing the Jewish paternal haplotypes(Very likely, Aran influence to – since Jews and Arabs share 40% of the same genetic material) in Black Ethiopians is from 2010, more recent than ANY of your data. The data in your last response i svery mixed, even contradicting itself in many places – yes, I know, SUPPOSEDLY written by TOP NOTCH experts, who, incidentally, have changed their terms many times so it appears they are say something, but meaning something else – ESPECIALLY, haplotype V, M(35), or whatever other term they use.
CN
You wrote…
“It is YOU who has a problem accepting the Ancient, and, Modern Egyptians were Arab Semitic White, NOT, Black Nubian – I present the reasoning why your information is WRONG, so you turn it into an ethnic thing – I abhor rape, but no getting around it – Black Ethiopian men raped Arab women – seem you overlooked that, and, yes, Arab men raped Black Ethiopian women – FACT. This explains the Semitic haplomarkers – Jews and Arabs share up to 40% of the same genetic material – that is why the presence of Semitic haplomarkers in the Black Ethiopian Jew – BOTH Arabs, and, Jews, NOT just Jews. It also explains the more Caucasian looks which YOU keep attempting to explain away as phenotypical variation – IT IS NOT – it is do to admixture as shown by the presence of Semitic White haplomarkers. I know, you want to keep denying this, attributing it to Black genetic variation. Would you do that with President Obama? Yes, I know, his White half is Indo European Aryan, NOT Arab Semitic – clea
rly, you can see, he is MIXED. Yes, this is NOT always possible. When a person drops below 25% Black heritage, they most often can pass as what ever else their other ethnic composition is – i.e. – White. Many of the MIXED in Sudan, pass themselves off as Arabs because of the caste system there.”
I never said that the “Black Jews” were not phenotyically ‘African’. In fact, are Afrasian, Afro-Asiatic and previously stated! You seem to think that this admixture ‘MAKES” them WHITE! Wrong!
http://muse.jhu.edu/journals/history_in_africa/v032/32.1keita.html
It is the height of arrogance – YOURS – to reject the study which confirms that haplotype V which is E-M81 and E-M183 as ‘Arabic’ or ‘Jewish’ as this is geo-political bull.
QUOTE:
“BIDIRECTIONAL clinal variation in Egypt for various p49a,f TaqI Y RFLP haplotypes (Table 1) has been suggested to be likely related to specific military campaigns during and after the Middle Kingdom (Lucotte and Mercier 2003a). The events considered to have brought together northern and southern populations having different Y genetic profiles are:
the Egyptian campaigns against and/or colonization of lower Nubia during the Middle and New Kingdoms (respectively primarily Dynasty XII, ca. 1991-1785 BCE, and Dynasty XVIII, beginning ca. 1490 BCE);
the Nubian conquest of Egypt by the Napatan kingdom that created Dynasty XXV (ca. 730-655 BCE), centered near the fourth cataract (in the Republic of the Sudan);
the conquest of Egypt during the Greco-Roman period by southern Europeans; and
the migration of Arabic-speaking peoples from the Near East, and
much later the Turks from Anatolia, both during the Islamic period. The first two of these have been suggested to explain the pattern of the three most common haplotypes: V, XI, and IV.”
“There is further evidence from a phylogeographic perspective for the BIOHISTORICAL AFRICANITY of haplotype V. Biallelic markers on the non-recombining portion of the Y chromosome define clades that can be associated with the TaqI p49a,f variants (see e.g. al-Zahery et al 2003, who present a kind of genetic Rosetta stone). Haplotype V is associated with the M35/215 subclade, as is XI (in Africa), and IV with the M2/PN1/M180 subclade, both of the YAP/M145/M213 cluster. These lineages (“subclades”) subsuming haplotypes V/XI and IV, are joined by a transition mutation: “most notably the PN2 transition . . . unites two high frequency sub-clades, defined by M2/PN1/M180 mutations in sub-Saharan Africa, and M35/215 in north and east Africa.” (Underhill et al. 2001:50). In one system of Y haplotype taxonomy, the subclades are in Group III (Bosch et al. 2001, Cruciani et. al. 2002). In another system of classification, these lineages are in haplogroup E (Hammer and Zegura 2002). The PN2 transition therefore defines a widespread clade. It is noteworthy that Group III is said to account for 73% of the variation in Africa (Underhill et al. 2001).”
“The modern population of E-M215 and E-M35 lineages are almost identical, and therefore by definition age estimates based on these two populations are also identical. E1b1b (E-M215) and its dominant sub-clade E1b1b1 (E-M35) are believed to have first appeared in East Africa about 22,400 years ago.”
Cruciani et al., Fulvio; La Fratta, Roberta; Santolamazza, Piero; Sellitto, Daniele; Pascone, Roberto; Moral, Pedro; Watson, Elizabeth; Guida, Valentina et al. (May 2004), “Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa” (PDF), American Journal of Human Genetics 74 (5): 1014–1022, doi:10.1086/386294, PMC 1181964, PMID 15042509;
http://en.wikipedia.org/wiki/American_Journal_of_Human_Genetics
Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out of Africa
“We explored the phylogeography of human Y-chromosomal haplogroup E3b analyzing 3,401 individuals from five continents.
Our data refine the phylogeny of the entire haplogroup, which appears as a collection of lineages with very different evolutionary histories, and reveal signatures of several distinct processes of migration and/or recurrent gene flow that occurred in Africa and western Eurasia over the past 25,000 years.
In Europe, the overall frequency pattern of haplogroup E-M78 does not support the hypothesis of a uniform spread of people from a single parental Near Eastern population.
The distribution of E-M81 chromosomes in Africa closely matches the present are of distribution of Berber-speaking populations on the continent, suggesting a close haplogroup-ETHNIC group parallelism.
:oops: E-M34 chromosomes were more likely introduced into Ethiopia from the Near East.
In conclusion, the present study shows that earlier work based on fewer Y-chromosome makers led to rather SIMPLE HISTORICAL INTERPRETATIONS and highlights the fact that MANY POPULATION-GENETIC ANALYSES ARE NOT ROBUST TO A POORLY RESOLVED PHYLOGENY.”
Okay, wrong again, as usual…
I never said that the Ancient Egyptians were not Nubians-which seems to be your point.
To be an ‘Egyptian’ was to have been part of the Mystery Schools which were rites of passage for royal children chosen for the priesthood. It was the Nubian who established “The Hereditary Priests and Priestesses. Priest = Su. Priestess = Sua. Am Su Nesi. Am Sua Nesi. These were those who spoke to and for the ancestors = Akh. Many of these lived in the city of Tukh (Tu-aat-Khu-An-Su) Khonsu. This is where the Boy King called Tut-Ankh-Amun was born and you can see how the language used to translate the words is the major flaw in how the texts are interpreted. It was the embrace of Arabic as a Semitic language which moved Afro-Asiatic, which included Semitic languages spoken in Eritrea and Ethiopia. The Nubians always spoke Nubian and today in the Arabic onslaught of acculturation a return to Nubian and a rejection of Arabic has begun.
You do realize that the concept of sacred speech or language is the ancient Medju-Netjer? It is in the Bible, in Genesis Chapter 11: 1-9:
1 And the whole earth was of one language, and of one speech.
2 And it came to pass, as they journeyed from the east, that they found a plain in the land of Shinar; and they dwelt there.
3 And they said one to another, Go to, let us make brick, and burn them thoroughly. And they had brick for stone, and slime had they for morter.
4 And they said, Go to, let us build us a city and a tower, whose top may reach unto heaven; and let us make us a name, lest we be scattered abroad upon the face of the whole earth.
5 And the LORD came down to see the city and the tower, which the children of men builded.
6 And the LORD said, Behold, the people is one, and they have all one language; and this they begin to do: and now nothing will be restrained from them, which they have imagined to do.
7 Go to, let us go down, and there confound their language, that they may not understand one another’s speech.
8 So the LORD scattered them abroad from thence upon the face of all the earth: and they left off to build the city.
9 Therefore is the name of it called Babel; because the LORD did there confound the language of all the earth: and from thence did the LORD scatter them abroad upon the face of all the earth.
Babel is false construct based again on Semitic, rather than Afro-Asiatic. Using the skill of the scribes of the time, i.e., the Chartummim, the Hebrews and obviously the Greeks learned from the so-called “Egyptians” like Manetho who curiously did not speak ‘Egyptian’ but was fluent in Greek! If you cannot speak the language you are translating how do you translate that language into Greek? Where is your ‘Arabic’ language in all of this history. The main reason the history of Africa and the Kingdoms of Africans is so easily rejected is because the literature in their languages, such as Medjewi, and if you do not see Medjuwi (Medjuwia) as in Medju (Medu) than it is because you do not want to.
Why? “Let us go down, and THERE confound their language, that they may not understand one another’s speech.”
“Therefore is the name of it called Babel; because the LORD did there confound the language of all the earth: and from thence did the LORD scatter them abroad upon the face of all the earth.”
Then – the Greeks. Today – the Arabs. When Nubian is spoken no longer, the history of this Kingdom will be written by foreigners and it would not surprise anyone this chapter in ancient civilization will be nothing but a footnote in the shadow of an Arab ‘Egypt” that never existed. Nubia, like nearly all African nations will be characterized as ‘emulators’ of the Egyptian rather than the hereditary originators and codifiers of the culture – all because they were not isolationists and the most adept European linguists cannot classify the language of the Eastern Sudanese (Isetem Sut-Nesi) much less translate the ancient texts – so we are told. Yet, the hundreds of papyri already translated tell us who Sut-Sen-Nesi was/is but refuse to connect the dots.
Rowan, Kirsty (2006). “Meroitic – an Afroasiatic language?” (PDF). SOAS Working Papers in Linguistics. pp. 14: 169–206. Retrieved 2008-11-15.
http://www.soas.ac.uk/linguistics/research/workingpapers/volume-14/file37822.pdf
Meroitic Newsletter (Paris: Académie des Inscriptions et Belles-Lettres, 1968)
Böhm, Gerhard : “Die Sprache der Aithiopen im Lande Kusch” in Beiträge zur Afrikanistik, 34, (Wien: 1988). ISBN 3-85043-047-2.
Rilly, Claude (March 2004) “The Linguistic Position of Meroitic”, Sudan Electronic Journal of
Archaeology and Anthropology. (2007) La langue du Royaume de Meroe. Paris: Champion.
Welsby, Derek A.: The Kingdom of Kush, (London: British Museum Press, 1996), 189-195, ISBN 071410986X.
:twisted: You can say anything…. And when the ancient lands have been once again flooded (Kajaba and Merowe dams) and all the evidence of their antiquity and glory is confined to another watery grave – like the Assuan High Dam achieved, the ‘Egyptians’ will have erased from their sight the bane of their existence and another historically African treasure will become so much mud kicked back into the faces of all Africans fighting to keep Nubia alive ! This is why African-Americans so vehemently reject ‘White Egypt” as this is the pattern of white-washing used by the powers and principalities of governments which do what they do because they can. Calling ancient ‘Egypt’ Arab is like calling the 17th Century Dutch Afrikaaner of South Africa aboriginal. It will always be a lie.
This is why Arabs were never ‘Egyptians’ since if the literature had been written by them—
If the bodies being dug up were of their ancestors put on display to exact tourist dollars for the morbid curiosity of Europeans…well…????? the Prophet Muhammad (PBU) comes to mind…
http://s4.zetaboards.com/Afro_Asian_mt_DNA/forum/3760613/
the Skeptic
It is YOU who has a problem accepting the Ancient, and, Modern Egyptians were Arab Semitic White, NOT, Black Nubian – I present the reasoning why your information is WRONG, so you turn it into an ethnic thing – I abhor rape, but no getting around it – Black Ethiopian men raped Arab women – seem you overlooked that, and, yes, Arab men raped Black Ethiopian women – FACT. This explains the Semitic haplomarkers – Jews and Arabs share up to 40% of the same genetic material – that is why the presence of Semitic haplomarkers in the Black Ethiopian Jew – BOTH Arabs, and, Jews, NOT just Jews. It also explains the more Caucasian looks which YOU keep attempting to explain away as phenotypical variation – IT IS NOT – it is do to admixture as shown by the presence of Semitic White haplomarkers. I know, you want to keep denying this, attributing it to Black genetic variation. Would you do that with President Obama? Yes, I know, his White half is Indo European Aryan, NOT Arab Semitic – clearly, you can see, he is MIXED. Yes, this is NOT always possible. When a person drops below 25% Black heritage, they most often can pass as what ever else their other ethnic composition is – i.e. – White. Many of the MIXED in Sudan, pass themselves off as Arabs because of the caste system there.
– in around 1500 B.C. as I mentioned earlier – the Ethiopian Empire swept across Egypt into Yemen(the Levant)- Mostly Arab – Jews living among them. The Arabs drove them BACK to Ethiopia. Again, Arabs also, either raped,or, took, Ethiopian wives.Liya Kebede is a modern day example, an Ethiopian model – her father is Arab Semitic White, her mother Black Ethiopian. Try again friend.
You really have a thing for using the imagery of Arabs RAPING African women. This is the second or third time you made this reference.
As I have said so often…
you can say anything about the “Black Jews” and it does not make what you say true.
Whether you or anyone thinks that Blacks have a right to practice Judaism and call themselves Jews is irrelevant. Judaism is a religion, just like Islam, and Christianity. All religions have a political agenda.
http://www.nytimes.com/2010/08/28/nyregion/28blackjews.html
“Everyone agrees that the numbers have grown, and they should be noticed,” said Jonathan D. Sarna of Brandeis University, a pre-eminent historian of American Jewry. “Once, there was a sense that ‘so-and-so looked Jewish.’ Today, because of conversion and intermarriage and patrilineal descent, that’s less and less true. The average synagogue looks more like America.”
You can say anything…
Israel is Gay
NON sense – Rlbl ORIGINATES in the ME – you are a late comer. The ME is MOSTLY Semitic White(Arabs, the closely related Jews, and other Semites(most gone)- has been since history. By your name – I really don’t believe you are Jewish, or you would know this.TURKS(Ottomans) are INDO EUROPEAN ARYAN WHITE, NOT, Semitic White( Arabs, and the closely related Jews(ethnically – NOT by religion(any ethnic group). The Middle East was NEVER INDO EUROPEAN ARYAN WHITE – the Europeans(including Turks) were invaders/ occupiers of ARAB lands – Arab armies drove them out by 1922, AFTER over 400 years of occupation – NOTE: the skeptic Egypt was ALREADY part of the Arab ME(65% Arab, Ancient, and, Modern Egyptians, NOT, the Black Nubians)since when the Turks ruled/occupied Egypt.
the Skeptic
The falsha have an Ethnic paternal limk to the Jews – King Solomon and his men visited Ethiopia. QUOTE,”In contrast, Ethiopian Jews (Beta Israel) and Indian Jews (Bene Israel and Cochini) cluster with neighbouring autochthonous populations in Ethiopia and western India, respectively, despite a clear paternal link between the Bene Israel and the Levant. These results cast light on the variegated genetic architecture of the Middle East, and trace the origins of most Jewish Diaspora communities to the Levant.” – translation- Jewish men had Black Ethiopian wives: The genome-wide structure of the Jewish people: http://www.nature.com/nature/journal/v466/n7303/abs/nature09103.html
– in around 1500 B.C. as I mentioned earlier – the Ethiopian Empire swept across Egypt into Yemen(the Levant)- Mostly Arab – Jews living among them. The Arabs drove them BACK to Ethiopia. Again, Arabs also, either raped,or, took, Ethiopian wives.Liya Kebede is a modern day example, an Ethiopian model – her father is Arab Semitic White, her mother Black Ethiopian. Try again friend.
NOTE: the skeptic Egypt was ALREADY part of the Arab ME(65% Arab, Ancient, and, Modern Egyptians, NOT, the Black Nubians)since when the Turks ruled/occupied Egypt.
Boo-Yah!
Re: Ma’at: Declarations of Innocence
Mon, June 14, 2010 – 1:51 PM
42 Declarations of Innocence or Admonitions of Maát
1. I have not committed transgression.
2. I have not committed robbery with violence.
3. I have not stolen.
4. I have not slain men and females (women).
5. I have not stolen grain.
6. I have not purloined offerings.
7. I have not stolen the property of the Goddess/God.
8. I have not uttered lies.
9. I have not carried away food.
10. I have not uttered curses.
11. I have not committed adultery, i have not lain with men. – Violated by both Greeks and Romans!
12. I have made none to weep.
13. I have not eaten the heart.
14. I have not attacked any female (woman)/man.
15. I am not a man of deceit.
16. I have not stolen cultivated land.
17. I have not been an eavesdropper.
18. I have not slandered [no female (woman)/man].
19. I have not been angry without just cause.
20. I have not debauched the wife/husband of any female (woman)/man.
21. I have not debauched the wife of [any] man.
22. I have not polluted myself.
23. I have terrorized none.
24. I have not transgressed [the law].
25. I have not been wroth.
26. I have not shut my ears to the words of truth.
27. I have not blasphemed.
28. I am not a man of violence.
29. I have not been a stirrer up of strife.
30. I have not acted with undue haste.
31. I have not pried into matters.
32. I have not multiplied my words in speaking.
33. I have wronged none, i have done no evil.
34. I have not worked witchcraft against the king.
35. I have never stopped [the flow of] water.
36. I have never raised my voice.
37. I have not cursed the Goddesses/God.
38. I have not acted with arrogance.
39. I have not stolen the bread of the goddesses/gods.
40. I have not carried away the khenfu cakes from the Spirits of the dead.
41. I have not snatched away the bread of the child, nor treated with contempt the god of my city.
42. I have not slain the cattle belonging to the goddess/god.
http://tribes.tribe.net/kmt/thread/cb385c77-7888-40bd-8f33-c325e7ce61af
Saying one is an ‘Egyptian’ once meant living according to the precepts. Instead he abandoned the ‘Old ways’ which apparently were only a cultural assimilation by the peoples of the Delta to South ‘Egypt,’ by way of the pre-dynastic Nubians – whose temples – ironically are in this watery grave called Lake Nasser!
What does it mean to be an ‘Egyptian’ today?
MODERN EGYPT : http://www.world66.com/africa/egypt/history
“The modern history of Egypt is marked by Egyptian attempts to achieve political independence first from the Ottoman Empire and then from the British. In the first half of the nineteenth century MUHAMMAD ALI AN ALBANIAN and the OTTOMAN VICEROY IN EGYPT attempted to create an Egyptian empire that extended to Syria and to REMOVE EGYPT FROM TURKISH CONTROL. Ultimately he was unsuccessful and true independence from FOREIGN POWERS would not be achieved until midway through the next century.
Foreign including British investment in Egypt and Britain’s need to maintain control over the Suez Canal resulted in the British occupation of Egypt in 1882. Although Egypt WAS GRANTED INDEPENDENCE IN 1922, British troops were allowed to remain in the country to safeguard the Suez Canal.
NOTE:
In 1952 the Free Officers led by Lieutenant Colonel Gamal Abdul Nasser took control of the government and removed King Faruk from power. In 1956 Nasser as Egyptian president announced the nationalization of the Suez Canal an action that resulted in the tripartite invasion by Britain France and Israel. Ultimately however Egypt prevailed and the last British troops were withdrawn from the country by the end of the year.”
SUCCESSION IN EGYPT THROUGH GENERALS IN EGYPTIAN ARMY
http://www.historylearningsite.co.uk/gamal_abdel_nasser.htm
“Under Neguib, civilian titles as associated with the Royal Family, were banned. Privileges associated with the ‘old way’ were also banned. Laws were brought in that limited the amount of land someone could own and they also widened the opportunities for land ownership. In 1961, Nasser nationalised a number of corporations so that the wealth that they generated could be used to improve the lifestyle of the Egyptian people. One year later, a decision was ANNOUNCED THAT EGYPT WOULD BE RUN ON ARAB SOCIALIST lines. During Nasser’s time in office, the Aswan High Dam was completed. This was a project that generated world-wide attention. However, iron and steel mills, aluminum plants, car and food factories were also built. In total, over 2000 new factories were built in Egypt in Nasser’s time.
However, Nasser suffered a major blow when Egypt and other Arab nations were beaten by Israel in the Six-Day War of 1967. By this year, Egypt was seen as the leading Arab nation and the Arab people looked to Egypt for leadership. For Nasser, the comprehensive defeat by Israel was a serious blow and he offered his resignation. This was rejected by the people who took to the streets in June 1967 to demonstrate their support for Nasser. After the war, Nasser went to great efforts to modernise the Egyptian military and this remained one of his primary aims until his death in September 1970. His death was followed by an outpouring of national grief in Egypt. Nasser was succeeded by Anwar Sadat.”
HIGH ASWAN DAM: Aswan Dam Documentary Part 1
http://www.youtube.com/watch?v=V0_90KxYD0Q
“Muhammad Ali Pasha al-Mas’ud ibn Agha (Arabic: محمد علي باشا, Mʋhɑm̑ɑd Oɑlí Báşá) (Mehmet Ali Pasha in Albanian; Kavalalı Mehmet Ali Paşa in Turkish)[2] (4 March 1769 – 2 August 1849) was an Albanian commander in the Ottoman army, who became Wāli, and self-declared Khedive of Egypt and Sudan. Though not a modern nationalist, he is regarded as the founder of modern Egypt because of the dramatic reforms in the military, economic and cultural spheres that he instituted. He also ruled Levantine territories outside Egypt. The dynasty that he established would rule Egypt and Sudan until the Egyptian Revolution of 1952.”
Albanian – not ‘Egyptian’ and yet, Muhammad Ali Pasha is regarded as “The Father of Modern Egypt.” Not an Arab. Not an Arab!
http://www.youtube.com/watch?v=5V3BSkCVsXg&NR=1
Y-DNA – Ethnic Group Distribution
http://en.wikipedia.org/wiki/Y-DNA_haplogroups_by_ethnic_groups
The High Dam – Assuan:
This dam violates an ancient precept, i.e, one of the 42 Declarations of Innocence:
35. i have never stopped [the flow of] water.
Declarations of Innocence:
http://tribes.tribe.net/kmt/thread/cb385c77-7888-40bd-8f33-c325e7ce61af
If Nasser were truly an Egyptian he would not have broken this precept or this one: