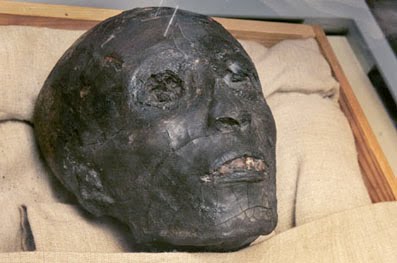
Despite the refusal of the Secretary General of the Egyptian Supreme Council of Antiquities, Zahi Hawass, to release any DNA results which might indicate the racial ancestry of Pharaoh Tutankhamen, the leaked results reveal that King Tut’s DNA is a 99.6 percent match with Western European Y chromosomes.
The DNA test results were inadvertently revealed on a Discovery Channel TV documentary filmed with Hawass’s permission — but it seems as if the Egyptian failed to spot the giveaway part of the documentary which revealed the test results.
Hawass previously announced that he would not release the racial DNA results of Egyptian mummies — obviously because he feared the consequences of such a revelation.
On the Discovery Channel broadcast, which can be seen on the Discovery Channel website here, or if they pull it, on YouTube here, at approximately 1:53 into the video, the camera pans over a printout of DNA test results from King Tut.
Firstly, here is a brief explanation of the results visible in the video. It is a list of what is called Short Tandem Repeats (STRs).
STRs are repeated DNA sequences which are “short repeat units” whose characteristics make them especially suitable for human identification.
These STR values for 17 markers visible in the video are as follows:
DYS 19 – 14 (? not clear)
DYS 385a – 11
DYS 385b – 14
DYS 389i – 13
DYS 389ii – 30
DYS 390 – 24
DYS 391 – 11
DYS 392 – 13
DYS 393 – 13
DYS 437 – 14 (? not clear)
DYS 438 – 12
DYS 439 – 10
DYS 448 – 19
DYS 456 – 15
DYS 458 – 16
DYS 635 – 23
YGATAH4 – 11
What does this mean? Fortunately, a genius by the name of Whit Athey provides the key to this list. Mr Athey is a retired physicist whose working career was primarily at the Food and Drug Administration where he was chief of one of the medical device labs.
Mr Athey received his doctorate in physics and biochemistry at Tufts University, and undergraduate (engineering) and masters (math) degrees at Auburn University. For several years during the 1980s, he also taught one course each semester in the electrical engineering department of the University of Maryland. Besides his interest in genetic genealogy, he is an amateur astronomer and has his own small observatory near his home in Brookeville, MD.
He also runs a very valuable website called the “Haplogroup Predictor” which allows users to input STR data and generate the haplogroup which marks those STR data.
For those who want to know what a haplogroup is, here is a “simple” definition: a haplogroup is a group of similar haplotypes that share a common ancestor with a single nucleotide polymorphism (SNP) mutation.
Still none the wiser? Damn these scientists.
Ok, let’s try it this way: a haplotype is a combination of multiple specific locations of a gene or DNA sequence on a chromosome.
Haplogroups are assigned letters of the alphabet, and refinements consist of additional number and letter combinations, for example R1b or R1b1. Y-chromosome and mitochondrial DNA haplogroups have different haplogroup designations. In essence, haplogroups give an inisight into ancestral origins dating back thousands of years.
By entering all the STR data inadvertently shown on the Discovery video, a 99.6 percent fit with the R1b haplogroup is revealed.
The significance is, of course, that R1b is the most common Y-chromosome haplogroup in Europe reaching its highest concentrations in Ireland, Scotland, western England and the European Atlantic seaboard — in other words, European through and through.
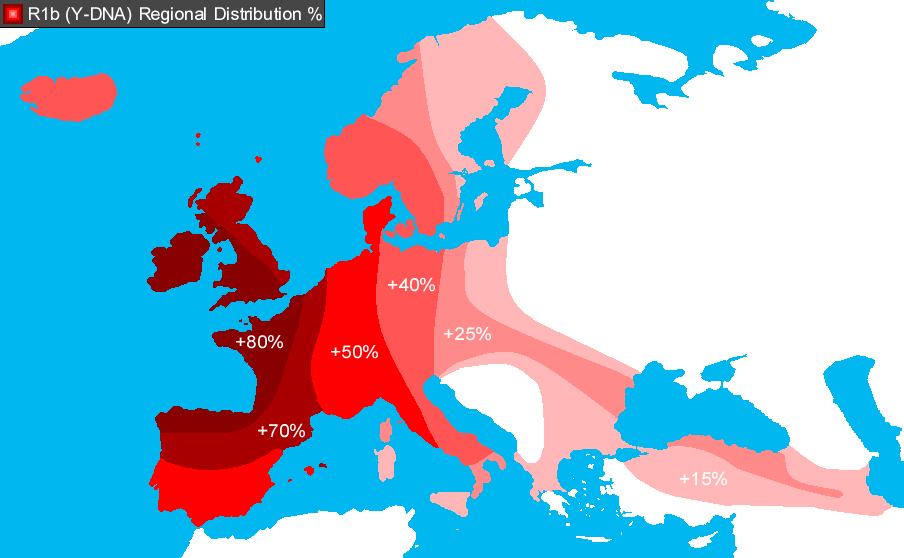
So much for the Afro-centrists and others who have derided the very obvious northwestern European appearance of a large number of the pharonic mummies. It seems like March of the Titans was right after all…
RELATED ARTICLES
- Researchers Warn Vaping is just as Harmful as Cigarettes
- DNA test to be added to Covid screening
- Justice in Corrupt Germany: 8 out of 9 Immigrants who Gang Raped Minor Girl in Park ON VIDEO, set Free
- FDA says Targeted Gene Therapy may CAUSE Cancer instead of Curing it
- Canada Confirms Undisclosed Presence of DNA Sequence in Pfizer Shot










No dude. You can cut out the facts you don’t like but Family Pedia tells the specifics you reject.
Mt-DNA Branching Structure:
L → L1 → L3 → L3M L3N → R, U, etc.
Factors of ‘African’ motif inheritance in order of haplogroup appearances:
16320C : L0a, L0a1d, L0a2c.
16320T : L0, M1a2b, M2b, M10a2b, M27a, R6a1a, K1c2.
Conversely, both Amhara and Tigrai U2 sequences share an HVS-I motif, 16051G, 16189C, 16234T, 16294T, clearly ‘African’ not ‘European’ motifs.
16051G : L0b, L1c1a1a1b, M9b, U2, U9a, G2a2, C1d, A7.
16189C : L2a1b, L2a1f, L2a1i, L2a1j, L2a2, L2d, L3b2, L3d3a, L3e2b, L3x2b, L4b1, M1, M2b, M7a1a2, M7b2, M7e, M8a2a, M13a, M29, M45, M47, M51, R11’b, R30b, R31b, U1a’c, U2d, U2e, U5b1b, U5b1c, U6a1, U6a2, U6a3, U6c, U8b, N9a5, N9b, A4b, A7, A9, A2e, A4b, C1b4, Z4, D4b2a2a, D5, F1b’d, G2a1c, G2a3, G3a2, H1b, H1f, T1, S3, X, Y1a1.
16189T : L2’3’4’6, D5c.
16189A : X2e1. If European CRS is 16189T.
16234T : L0a2c, U2, U4a1c, U8b, K1a1b1a, K1a4b, K1a9, K1a8, B4d3, B5b2, G2a1a.
16294C : L1c1c. If European CRS 16294C. L1c1c is the at the root of L0 – L1 clearly ‘African’ (“Negroid”), therefore this is inherited by ‘Caucasians’ tracing this particular lineage back to Africa.
16294T : L1c, L2a, M29b, M40a, M51, N9a6a, N9b2, P1b, U2d.
You can say anything. The results argue against what you say…Didn’t you know the devil is in the details….
All eight Ethiopian U6 samples descend from the major U6a1 founder, which spread from the Near East to northwestern Africa at appreciable frequencies (Maca-Meyer et al. 2003). Their absence in Yemen suggests that these U6 lineages have likely penetrated to Ethiopia from the north rather than by the sea route from Arabia.
Conversely, both Amhara and Tigrai U2 sequences share an HVS-I motif, 16051-16189-16234-16294, that has not been sampled, to date, in North Africa but that can be found at low frequencies in populations of western Asia and the Caucasus (authors’ unpublished data).” To determine whether Caucasian the CRS values are 16051A, 16189T, 16234C,16294C. Otherwise, all other markers are South Asian and African, where African is characterized as Tigrai, clearly not ‘European” or Europoid.
If Caucasian : U2 subclade European Defining mutations : 152*, 1811, 15907, 16051, 16129*
http://www.eupedia.com/europe/european_mtdna_haplogroups.shtml
Nope. U is a multi-ethnic haplogroup.
“In human genetics, Haplogroup U is a human mitochondrial DNA (mtDNA) haplogroup, a group of people who descend from a woman in the Haplogroup R branch of the Genographic tree, who lived around 50,000 years ago. Her descendants gave birth to several different subgroups, some of which exhibit specific geographic homelands. The old age has led to a wide distribution of the descendant subgroups that harbor specific European, northern African, Indian, Arab, northern Caucasus Mountains and the Near East clades.[1]”
So try that again….”The old age has led to a wide distribution of the descendant subgroups that harbor SPECIFIC European, northern AFRICAN, INDIAN, ARAB, northern Caucasus Mountains and the NEAR EAST clades.”
http://familypedia.wikia.com/wiki/Haplogroup_U_%28mtDNA%29
The game is over.
Modern Tigrai – Ancient Kerma Mt-DNA ‘U’ Inheritance 16189C:
“The Tigrai T1a sequence matches a Kerma sequence from Nubia (Krings et al. 1999), whereas the Amhara T1b sequence shows a mutation at np 16320 on top of the common founder haplotype in the Near East (Richards et al. 2000).”
“All eight Ethiopian U6 samples descend from the major U6a1 founder, which spread from the Near East to northwestern Africa at appreciable frequencies (Maca-Meyer et al. 2003).
… Their ABSENCE in YEMEN suggests that these U6 LINEAGES have likely penetrated to Ethiopia from the north rather than by the sea route from Arabia.
… Conversely, both Amhara and Tigrai U2 sequences share an HVS-I MOTIF, 16051-16189-16234-16294, that has NOT been SAMPLED, to date, in NORTH AFRICA but that can be found at low frequencies in populations of WESTERN ASIA and the Caucasus (authors’ unpublished data).”
Western Asia:
…the consensus of sequence of U4 in EUROPE DIFFERS from the ROOT HAPLOTYPE of U by NINE SUBSTITUTIONS. One Tigrai and one Yemeni U sample shared a transition at np 6386 that defines U9, which was recently detected in South Asia (Quintana-Murci et al. 2004). Therefore, haplogroups U4 and U9 are SISTER clades within a CLADE that is defined by one control-and one CODING-REGION MUTATION.”
It is not necessary to PERMIT YOU to translate for anyone – the authors have been very clear in stating that U us multi-ethnic specific and not as you put it ‘Caucasian’.
Background
http://www.familytreedna.com/public/U6mtdna/default.aspx
U6 haplogroup mitochondrial DNA project – Background
“The U6 haplogroup has a southwestern asia origin approximately 40,000 or 45,000 years ago. The early Upper Paleolithic carrying U6 return to Africa from the Mediterraneen area. This expansion had affected, not only north Africa, but also Near East, Iberian Peninsula and Canary Island. Then U6 haplogroup diffused to Europe through Gibraltar straits or Sicily, but also in sub-saharan regions.”
You do not have to like it but the truth shall make you free.
U jelly black supremacist? :-D
No worries,you still have Ethiopia.They had a descent country 3000 years ago.Sure it was nothing special,but it’s still better than nothing :wink:
the Skeptic
Taken from YOUR link – QUOTE,”U6 is thought to have entered North Africa around 30,000 years ago from the Near-East(*** as in Middle East – Arab Semitic White*** – MY EMPHASIS). In spite of the highest diversity of Iberian U6, Maca-Meyer argues for an Iberian origin of this clade based on the highest diversity of subclade U6a in that region,[17] where it would have arrived from West Asia. She estimates the age of U6 between 25,000 and 66,000 years BP. However U6 has its highest frequencies in North Africa and seems to be a specific haplogroup of that region.
U6 has three main subclades:[17]”
From your article – QUOTE,” “…All eight Ethiopian U6 samples descend from the major U6a1 founder, which spread from the Near East to northwestern Africa at appreciable frequencies (Maca-Meyer et al. 2003). Their absence in Yemen suggests that these U6 lineages have likely penetrated to Ethiopia from the north rather than by the sea route from Arabia. Conversely, both Amhara and Tigrai U2 sequences share an HVS-I motif, 16051-16189-16234-16294, that has not been sampled, to date, in North Africa but that can be found at low frequencies in populations of western Asia and the Caucasus (authors’ unpublished data).” To determine whether Caucasian the CRS values are 16051A, 16189T, 16234C,16294C. Otherwise, all other markers are South Asian and African, where African is characterized as Tigrai, clearly not ‘European” or Europoid.
Haplogroup U : http://isical.academia.edu/vasuluTS/Papers/412863/Mitochondrial_DNA_distribution_of_Macohaplogroup_N_and_its_haplogroup_U_and_subhaplogroup_R_in_Indian_populations
“…Despite the fact that haplogroup-N lineages occur at equally high frequency in Ethiopians and Yemenis, only three haplotypes (representing 3% of the total Yemeni sample) were found to match between the two populations. This suggests that the immediate source populations for these lineages in the Near East, from which they derive, could have been different.”
“Though present-day Ethiopia is a land of great ethnic diversity, the majority of Ethiopians speak different Semitic, Cushitic, and Omotic language that belong to the Afro-Asiatic linguistic phylum.
“…Maternal lineages of Semitic – (Amharic, Tigrinya, and Gurage) and Cushitic – (Oromo and Afar) speaking populations studied here reveal that their mtDNA pool is a nearly equal composite of sub-Saharan African and western Eurasian lineages. This finding, consistent with classic genetic-marker studies (Cavalli-Sforz 1997) and previous mtDNA results, is also in agreement with a similarly high proportion of Western Asian Y chromosomes in Ethiopians (Passarino et al. 1998; Semino et al. 2002), which supports the view (Richards et al. 2003) that the observed admixture between sub-Saharan Africans and, most probably, western Asian ancestors of the Ethiopian populations applies to their gene pool in general.
…On the other hand, significant differences in the proportions of derived lineages of haplogroup N between northwestern and south-central samples from Ethiopia are consistent with the proximity of the Tigrinya region (Aksum) and Eritrea to the coast of the Red Sea, the latter having mediated gene flow with Egypt and southern Arabia – perhaps, in particular with the rise of Semitic cultural influence in the region.
…In contrast, the similarity of Amharas and Oromos, also expressed in other genetic loci (Fort et al. 1998; Corbo et al. 1999), supports the idea that “amharization” may have been largely a sociocultural rather than a genetic phenomenon. Yet, it is important to add here that Y-chromosomal haplogroup J1-M267, which is widespread throughout Arab-speaking countries and encompasses a third of Amharan Y chromosomes, has hardly penetrated the Cushitic-speaking Oromo population (Semino et al. 2004).””
Permit me to translate for you – Southwest and West Asia ARE THE MIDDLE EAST(formerly, NEAR EAST) – Arab Semitic White – the haplomarkers spread from Yemen and Arabia through North Africa. AGAIN haplomarker U1-8 are Caucasian haplomarkers – Arabs are NOT Indo European Aryan White(hence, NOT European, but Middle Eastern(Semitic) Whites).
Try again.
CN
“Just when I thought I was out they pull me back in.” – Michael Corleone
ScienPapers/Ethiopian Mitochondria DNA
http://docs.google.com/viewer?a=v&q=cache:TfB_Iyv8mVIJ:www.africandna.com/ScienPapers%255CEthiopian_Mitochondrial_DNA_Heritage.pdf+16189C+motif+%2B+mtdna+%2B+African&hl=en&gl=us&pid=bl&srcid=ADGEEShRD6xF78NFAa11Bj03kYsjFtsRncbUm4kwiV8wnz8mGot9PkHNgb6Q_1ha52myZzHzxoBEoX2uZKcfWlOC9V9kMOiTeS2tCul4Y2sDLdQEWPACgc6nF2gdS8NUhzmOlHr7KNkf&sig=AHIEtbQulGYe-r8u-9JtVjIY9Yv4400vRw
Excerpt: Google Books p.764 – 765
Modern Tigrai – Ancient Kerma Mt-DNA ‘U’ Inheritance 16189C:
“The Tigrai T1a sequence matches a Kerma sequence from Nubia (Krings et al. 1999), whereas the Amhara T1b sequence shows a mutation at np 16320 on top of the common founder haplotype in the Near East (Richards et al. 2000).” South Asia.
“Five of the six T2 sequences detected among Amhara and Tigrai samples shared a transition at np 16292 that is widespread in the haplogroup T context in Europe, the Near East, and North Africa. However, two Tigrai T2 sequences share a combination of four downstream HVS-I mutations that have not been reported elsewhere.”
“…N1a is a minor mtDNA haplogroup that has been observed at marginal frequencies in European, Near Eastern, and Indian populations (Mountain et al. 1995; Richards et al. 2000). It occurs at a significant frequency in both Ethiopian and Yemeni populations.”
“…Six Ethiopian N1a lineages, restricted to Semitic-speaking subpopulations, show low haplotype diversity and include an exact HVS-I sequence match with a published N1a sequence from Egypt (Krings et al. 1999). A related sequence, from southern Sudan (Krings et al. 1999) was misclassified as a member of the L1a clade (Salas et al. 2002).”
“Yemeni N1a sequences, on the other hand, display a high level of haplotype (h=0.89) and nucleotide (p-2.75+/-1) diversity, combined with the highest frequency (6.9%) of this haplogroup reported so far.”
“…Two Ethiopian haplogroup X sequences from this study have been characterized elsewhere as belonging to North and East African-specific subclade X1 (Reidla et al. 2003). A control-region sequence similar to the Tigrai X1 haplogroup was found recently in a Gurna sample from Egypt, though it was probably mislabled as “L3” by the authors, since no coding-region markers specific to either haplogroup X or L3 were determined in that study (Stevanovitch et al. 2004). Both Yemeni X sequences, in contrast, belong to the major western Eurasian subclade X2.” (Not European !)
“…Haplogroup –U4 lineages are spread at moderate frequencies all over Europe, western Siberia, and south-western Asia and coalesce to their most recent common ancestor within the Middle or Late Upper Paleolithic period (Richards et al. 2000; Tambets et al. 2003). Upper Paleolithic –
On the basis of complete sequence data (Finnila et al. 2001; Herrnstadt et al. 2002),
…the consensus of sequence of U4 in Europe differs from the root haplotype of U by nine substitutions. One Tigrai and one Yemeni U sample shared a transition at np 6386 that defines U9, which was recently detected in South Asia (Quintana-Murci et al. 2004). Therefore, haplogroups U4 and U9 are sister clades within a clade that is defined by one control-and one coding-region mutation.”
“…All eight Ethiopian U6 samples descend from the major U6a1 founder, which spread from the Near East to northwestern Africa at appreciable frequencies (Maca-Meyer et al. 2003). Their absence in Yemen suggests that these U6 lineages have likely penetrated to Ethiopia from the north rather than by the sea route from Arabia. Conversely, both Amhara and Tigrai U2 sequences share an HVS-I motif, 16051-16189-16234-16294, that has not been sampled, to date, in North Africa but that can be found at low frequencies in populations of western Asia and the Caucasus (authors’ unpublished data).” To determine whether Caucasian the CRS values are 16051A, 16189T, 16234C,16294C. Otherwise, all other markers are South Asian and African, where African is characterized as Tigrai, clearly not ‘European” or Europoid.
Haplogroup U : http://isical.academia.edu/vasuluTS/Papers/412863/Mitochondrial_DNA_distribution_of_Macohaplogroup_N_and_its_haplogroup_U_and_subhaplogroup_R_in_Indian_populations
“…Despite the fact that haplogroup-N lineages occur at equally high frequency in Ethiopians and Yemenis, only three haplotypes (representing 3% of the total Yemeni sample) were found to match between the two populations. This suggests that the immediate source populations for these lineages in the Near East, from which they derive, could have been different.”
“Though present-day Ethiopia is a land of great ethnic diversity, the majority of Ethiopians speak different Semitic, Cushitic, and Omotic language that belong to the Afro-Asiatic linguistic phylum.
“…Maternal lineages of Semitic – (Amharic, Tigrinya, and Gurage) and Cushitic – (Oromo and Afar) speaking populations studied here reveal that their mtDNA pool is a nearly equal composite of sub-Saharan African and western Eurasian lineages. This finding, consistent with classic genetic-marker studies (Cavalli-Sforz 1997) and previous mtDNA results, is also in agreement with a similarly high proportion of Western Asian Y chromosomes in Ethiopians (Passarino et al. 1998; Semino et al. 2002), which supports the view (Richards et al. 2003) that the observed admixture between sub-Saharan Africans and, most probably, western Asian ancestors of the Ethiopian populations applies to their gene pool in general.
…On the other hand, significant differences in the proportions of derived lineages of haplogroup N between northwestern and south-central samples from Ethiopia are consistent with the proximity of the Tigrinya region (Aksum) and Eritrea to the coast of the Red Sea, the latter having mediated gene flow with Egypt and southern Arabia – perhaps, in particular with the rise of Semitic cultural influence in the region.
…In contrast, the similarity of Amharas and Oromos, also expressed in other genetic loci (Fort et al. 1998; Corbo et al. 1999), supports the idea that “amharization” may have been largely a sociocultural rather than a genetic phenomenon. Yet, it is important to add here that Y-chromosomal haplogroup J1-M267, which is widespread throughout Arab-speaking countries and encompasses a third of Amharan Y chromosomes, has hardly penetrated the Cushitic-speaking Oromo population (Semino et al. 2004).”
In other words, it is false to claim that all mt-DNA haplotypes U, T, N, P, X, Y are ‘Caucasian’ when clearly there are markers specific to ethnic groups which is totally different than all are White-Semitic….blah, blah, blah…
Mt-DNA Branching Structure:
L → L1 → L3 → L3M L3N → R, U, etc.
Factors of ‘African’ motif inheritance in order of haplogroup appearances:
16320C : L0a, L0a1d, L0a2c.
16320T : L0, M1a2b, M2b, M10a2b, M27a, R6a1a, K1c2.
Conversely, both Amhara and Tigrai U2 sequences share an HVS-I motif, 16051G, 16189C, 16234T, 16294T, clearly ‘African’ not ‘European’ motifs.
16051G : L0b, L1c1a1a1b, M9b, U2, U9a, G2a2, C1d, A7.
16189C : L2a1b, L2a1f, L2a1i, L2a1j, L2a2, L2d, L3b2, L3d3a, L3e2b, L3x2b, L4b1, M1, M2b, M7a1a2, M7b2, M7e, M8a2a, M13a, M29, M45, M47, M51, R11’b, R30b, R31b, U1a’c, U2d, U2e, U5b1b, U5b1c, U6a1, U6a2, U6a3, U6c, U8b, N9a5, N9b, A4b, A7, A9, A2e, A4b, C1b4, Z4, D4b2a2a, D5, F1b’d, G2a1c, G2a3, G3a2, H1b, H1f, T1, S3, X, Y1a1.
16189T : L2’3’4’6, D5c,
16189A : X2e1
16234T : L0a2c, U2, U4a1c, U8b, K1a1b1a, K1a4b, K1a9, K1a8, B4d3, B5b2, G2a1a.
16294C : L1c1c.
16294T : L1c, L2a, M29b, M40a, M51, N9a6a, N9b2, P1b, U2d.
mtDNA Haplogroup predicting;
http://docs.google.com/viewer?a=v&q=cache:u0ohYZbRuTwJ:volgagermanbrit.us/documents/mtDNA_Haplogroups.pdf+mtdna+%2B+16189C&hl=en&gl=us&pid=bl&srcid=ADGEESiOKg3hdfV8c_Cg1-ceybparp0nuBSIw_cb2Z7H8Ga5Z_hWJmtTf6SzQ5tbz7mekoI4m9hw5YTYDIvs6umHNSPpRogyTR8xI8O5toMjmbuifNiCHiR7AS1ViL861Hdp4-kQameV&sig=AHIEtbRG3_qrlExMRtvSULL1peg3hXVRng
In other words the ethnicity, i.e., ‘race’ is based on the evolution of natural selection motifs already determined as an ‘African’ (“Negroid”) marker, which inherited markers are dated to YBP according to the mutation rates which determine the derivation of the new haplogroup. It is fair to suggest that the phenotypes of the new haplogroups and clades within clades would not significantly change and the Near Eastern origination is equated with Asian creating the Afro-Asiatic man, who is more African regardless of lighter complexion and straighter hair.
Compare Cambridge Reference Sequence (CRS) for so-called European polymorphisms. Genetics shows the genotype-phenotype of the ‘African’ was first and very diverse. Therefore, it is always incorrect to state that Africans received genes from Caucasians, since the latter group was last on the genetic ladder.
16051A, 16189T, 16234C, 16294C, 16320C. 16129G, 16216A, 16311T, 16172T, 16223C, 16519T, 16183A, 16311T, 16292C, 6389C.
U9 : 3531A, 3834A, 6386T, 14094C
“There are altogether around 80 different ethnic groups in Ethiopia today, with the two largest being the Oromo and the Amhara, both of which speak Afro-Asiatic languages.”
Mt-DNA Kerma –
“The Tigrai T1a sequence matches a Kerma sequence from Nubia (Krings et al. 1999), whereas the Amhara T1b sequence shows a mutation at np 16320 on top of the common founder haplotype in the Near East (Richards et al. 2000).”
Kerma:
“The development of Kerma was contemporaneous with the C-Group in Lower Nubia. While Egypt was concerning itself with the control of Lower Nubia and the C-Group, Kerma was slowly developing its trade and culture beyond this buffer zone. There were three periods of the Kerma culture – Old Kerma, Middle Kerma and Classic Kerma. By c.1650 BC, Kerma had become densely populated and controlled a centralized state stretching from at least the First to the Fourth Cataracts. It was during the mid to latter part of the Second Intermediate Period of Egypt and during the Classic Period of Kerma that saw the apex of its wealth and power. Kerma was sacked in c.1500 BC, when the whole region became part of the Egyptian New Kingdom empire.”
http://www.theancientegyptians.com/Kerma.htm
CN, you wrote…
“You also attempted to claim haplomarker U(1-8) included Blacks – term Africans was used to to describe this haplomarker and subclades -, which, perhaps, confused you – better term would have been African White – haplomarker U is a CAUCASIAN haplomarker – the only exception is haplomarker U9 – WHICH YOU DID NOT TOUCH ON – occurs in 2-3% of the Black Ethiopians.Try again, friend.”
Nope. Not Caucasian – Asian…and if you really want to get an idea of the Asian phenotype…
mt-DNA at all but South Asian. These Blacks are Afro-Asiatics!
Looks to me that you were had.
The Skeptic
To summarize for you – King Tut’s DNA is White, Semitic – R1b1 originates in the Middle East, NOT in Europe. The Ancient and Modern Egyptians are Arab Semitic White, NOT, Black Nubians – the two culures/ethnic groups did have a large amount of mixing(up to 30%). NOME of your refer to Africans as “Black” – Sub Saharan and/or Nilotic is the term used for Blacks – your BIGGEST problem is, you want to call African, Blacks – the humanoids, that all humans are descended from,INCLUDING Blacks,are descended from, were NOT classified as Black. North Africa is Arab Semitic White. The Middle East was formerly called the Near East, West Asia, South West Asia. Once you comprehend this, we can have an intelligent discussion. Egypt, starting with Lower (Northern) Egypt, is predominantly Arab Semitic White – has been since the building of the Pyramids – as you cross midway, there is Arab/Black mixes(up to 30%), 5% all other ethnic groups. Heading into the smaller far southwest(part of Upper (Southern)Egypt) corner – formerly Nubia/Sudan/Kemit – annexed by the Ancient Egyptians in 1521 B.C., Egypt becomes mostly Black. Yes, you presented skull studies from this region to back your claim, the Ancient Egyptians wer Black(NOT TRUE)- take it from the Northern part of Egypt – they are arab semitic White. You also attempted to claim haplomarker U(1-8) included Blacks – term Africans was used to to describe this haplomarker and subclades -, which, perhaps, confused you – better term would have been African White – haplomarker U is a CAUCASIAN haplomarker – the only exception is haplomarker U9 – WHICH YOU DID NOT TOUCH ON – occurs in 2-3% of the Black Ethiopians.Try again, friend.
White – Semitic – Arabs…whitesemiticarabswhichsemiticarabs…ugh!
In them old cotton fields back home.
Did they have the Bol weevil back then?
the Skeptic
You really need to accept the Ancient Egyptians were Arab Semitic White, NOT the Black Nubians – your BIGGEST error is claiming Africans are Black – NOT what anything YOU quoted, said – when YOUR experts refer to Blacks – they are called sub Saharan, Nubians, Nilotics, NOT Africans – when humanoids are discovered in Africa, they are Africans, NOT, Black – also – North Africa is Arab Semitic White. You also have a BAD habit of misinterpreting data, because you do NOT comprehend what you copy, and, paste. Also, west, South West, Near East, are all terms for Middle East as in Arab Semitic White. Once you get all this STRAIGHT, we can have a descent discussion – until next time.
CN
I am sure you will be pleased to know that I have grown bored with this back and forth between us, and will make my exit right here. I will visit to read every now and again. You really have to get over the wheel and the mantra…White-Semitic-Arabs…white-semitic.arabs…LOL.
the Skeptic –
You are NOT comprehending what you are copying/pasting – QUOTE, “Derivative clade U6a1 signals a POSTERIOR movement FROM East AFRICA BACK(MY EMPHASIS) to the Maghrib AND the NEAR EAST.” Near East, and, West Asia,is MIDDLES EAST as in Arab Semitic White – BACK – refers to BACK FLOW – returning to its ORIGINS. North Africa is ARAB SEMITIC WHITER – BERBERS(and MOORS) are mainly of Arab Semitic White heritage, 8-15% Black heritage, and, small amounts of Jewish Semitic, and, Indo European Aryan Whites. The Iberian Peninsula is in EUROPE – Indo European Aryan White.NO WHERE in any of the links YOU provided, does it refer to Africans as Black – that is YOUR insert/interpetation – Haplomarker U1-8 refers to CAUCASIANS ONLY haplomarkers – all regions discussed are Arab Semitic White, Indo European Aryan White, or BOTH – show me anytghing that referes to BLACK, NOT African – sorry buddy – that is ONLY YOUR interpetation – NOT as quoted -. Your sources do mention some haplomarkers U^ did EXPAND into East, and, West Africa – from Arab migrations- Near EastWest Asia = Middle East – this link will clarify things for you: http://en.wikipedia.org/wiki/Haplogroup_U_%28mtDNA%29
The links do NOT mention haplomarker U9 (which IS a Black haplomarker, occurring in low frequencies – 2-3% in Ethiopians)
Finally,
African (Black) Specific Clades according to mutation:
U6a and U6b share a common basal mutation (16219) that is not present in U6c.
U6b: shows a more patched and western distribution. In the Iberian peninsula U6b is more frequent in the North (while U6a is in the South). It has also been found in low amounts in Morocco, Algeria, Senegal and Nigeria. Estimated age: 24,500 – 8,000 BP. It has one subclade:
U6b1: found only in the Canary Islands and in the Iberian peninsula. Estimated age: c. 6000 BP.
U6c: only found in Morocco and Canary Islands. Estimated age: 6,000-17,500 BP.
U6a and U6b share a common basal mutation (16219) that is not present in U6c.
“A gene is essentially a sentence made up of the bases A, T, G, and C that describes how to make a protein. Any changes to those instructions can alter the gene’s meaning and change the protein that is made, or how or when a cell makes that protein. There are many different ways to alter a gene, just as there are many different ways to introduce typos into a sentence.”
16129A>G, 16129G>A : Traces 16129 back to ‘African’ specific mt-DNA L ancestral mutations not the L3 from which the so-called Eurasian mt-DNA M N R H etc., are supposed to be derived.
Mutation 16129A: Traces back to L2b, L2d, L3flb1, M1, M10a1, M21ab, M28a, M47, M5, M5a2, M7b1d, N9a1’3, N9b1b, A5c, A9, B4ada1a, B5a2, C4a1, Z1, D2, D4a, G2b2, F1a’c, G1b.
Mutation 16129G: Traces back to L0a1c, L0a2, L0a2d, L0d1c, L0f2b, L0k1, L1b, M1a5.
Mt-DNA L2b : Several “L2 haplotypes observed in Guineans and other West Africa populations shared genetic matches with East Africa and North Africa.[5] An origin for L2b, L2c, L2d and L2e in West or Central Africa seems likely.[4] The early diversity of L2 can be observed all over the African Continent, but as we can see in Subclades section below, the highest diversity is found in West Africa. Most of subclades are largely confined to West and western-Central Africa, it is therefore likely that the L2 first emerged in this African region.[6]”
Mt-DNA L0a1c : “Haplogroup L0a is most prevalent in South-East African populations (25% in Mozambique).[3] Among Guineans, it has a frequency between 1% and 5%, with the Balanta group showing increased frequency of about 11%. Haplogroup L0a has a Paleolithic time depth of about 33,000 years and likely reached Guinea between 10,000 and 4,000 years ago. It also is often seen in the Mbuti and Biaka-pygmies.”
In other words, U6 was derived from ‘African gene mutations’ making the difference between mt-DNA L to U6 only a matter of historical migration times.
This means that the African L0 gave rise to the African U6*. These ‘Berbers’ are ‘African’.
None of this genetics is as simple as some who play with it think. None.
U6 has three main subclades:
Subgroup U6a reflects the first AFRICAN expansion from the Maghrib returning to the east in Paleolithic times. Derivative clade U6a1 signals a POSTERIOR movement FROM East AFRICA BACK to the Maghrib AND the NEAR EAST.
TAKE YOUR PICK: Posterior may refer to:
(1) Posterior (anatomy), the end of an organism opposite to its head.
(2) Buttocks, as a euphemism
(3) Posterior probability, the conditional probability that is assigned when the relevant evidence is taken into account.
(4) Posterior tense, a relative future tense.
Derivative clade U6a1 signals a POSTERIOR movement FROM East AFRICA BACK to the Maghrib AND the NEAR EAST.
…This migration coincides with the probable Afroasiatic linguistic expansion. U6b and U6c clades, restricted to West Africa, had more localized expansions. U6b probably reached the Iberian Peninsula during the Capsian diffusion in North Africa.
…Two autochthonous derivatives of these clades (U6b1 and U6c1) indicate the arrival of North African settlers to the Canarian Archipelago in prehistoric times, most probably due to the Saharan desiccation. The absence of these Canarian lineages nowadays in Africa suggests important demographic movements in the western area of this Continent.
—Maca-Meyer 2003
U6a: It is the most widespread (from Canary Islands and Iberian Peninsula to Syria, Ethiopia and Kenya) and has HIGHEST DIVERSITY in EASTERN Africa. Estimated age: 24-27,500 BP. It has one major subclade:
U6a1: with similar distribution to U6a. Estimated age: 15-20,000 BP. It was also found in Brittany (France) at 4.5% frequency (1/22)[18]
Quote: autochthonous = aboriginal. North Africa INCLUDES up to East Sudan/Ethiopia:
“Two autochthonous derivatives of these clades (U6b1 and U6c1) indicate the arrival of North African settlers to the Canarian Archipelago in prehistoric times, most probably due to the Saharan desiccation. The absence of these Canarian lineages nowadays in Africa suggests important demographic movements in the western area of this Continent.”
NOTE : Mt-DNA U6 came before Mt-DNA U8.
CN
Thanks for the opening….
Haplogroup U is subdivided into Haplogroups U1-U8.
Haplogroup K is a subclade of U8
{my insert : Near East – West Asian CLADE
Haplogroup U8 – U8a: The Basques have the most ancestral phylogeny in Europe for the mitochondrial haplogroup U8a, a rare subgroup of U8, placing the Basque origin of this lineage in the Upper Palaeolithic. The lack of U8a lineages in Africa suggests that their ancestors may have originated from West Asia.[2]
U8b: This clade has been found in Italy and Jordan.[2]
Haplogroup K – (named ‘Katrine’ by Bryan Sykes) makes up a sizeable fraction of European and West Asian mtDNA lineages. It is now known it is actually a subclade of haplogroup U8b’K,[2] and is believed to have first arisen in northeastern Italy. Haplogroup UK shows some evidence of being highly protective against AIDS progression.[13].}
The old age has led to a wide distribution of the descendant subgroups that harbor SPECIFIC EUROPEAN, Berber, Indian, African, Arab, northern Caucasus Mountains AND the NEAR EAST CLADES.
http://en.wikipedia.org/wiki/Haplogroup_U_%28mtDNA%29
The Skeptic
FROM YOUR LINK- Quote,”U6 is thought to have entered North Africa around 30,000 years ago from the Near-East(***** NOW, called the Middle East as in Arab Semitic White***** – MY EMPHASIS). In spite of the highest diversity of Iberian U6, Maca-Meyer argues for an Iberian origin of this clade based on the highest diversity of subclade U6a in that region,[17] where it would have arrived from West Asia. She estimates the age of U6 between 25,000 and 66,000 years BP. However U6 has its highest frequencies in North Africa and seems to be a specific haplogroup of that region.”
Translation – Arabs(Semitic Whites) back flowed into Africa,and, mated with the Black population. Again – all U haplomarkers,and, subclades, EXCEPT U9(about 2-3% in Ethiopians – YOUR ARTICLE DID NOT MENTION THIS ONE AT ALL) are Caucasian haplomarkers. AND, AGAIN, Africans is USED, NOT Blacks.
CN
So in other words you disregard this cladistic specificity:
“Haplogroup U is subdivided into Haplogroups U1-U8. Haplogroup K is a subclade of U8.[2] The old age has led to a wide distribution of the descendant subgroups that harbor SPECIFIC European, Berber, Indian, African, Arab, northern Caucasus Mountains and the Near East CLADES.[3]
http://en.wikipedia.org/wiki/Haplogroup_U_%28mtDNA%29
the Skeptic
All haplomarker U and subclades, EXCEPT haplomarker U9(very small amounts in Ethiopians) is Caucasian.
CN
I have been to several other forums which deal with the haplogroup of King Tut. I was wondering whether we got it all wrong by not thinking of the mt-DNA U:
“U6: It is common in North Africa (around 10% of the people) [32] with a maximum of 29% in Algerian Berbers[33]
U6 has three main subclades:[17]
Subgroup U6a reflects the first African expansion from the Maghrib returning to the east in Paleolithic times.
…Derivative clade U6a1 signals a posterior movement from East Africa back to the Maghrib and the Near East. This migration coincides with the probable Afroasiatic linguistic expansion. U6b and U6c clades, restricted to West Africa, had more localized expansions.
…U6b probably reached the Iberian Peninsula during the Capsian diffusion in North Africa. Two autochthonous derivatives of these clades (U6b1 and U6c1) indicate the arrival of North African settlers to the Canarian Archipelago in prehistoric times, most probably due to the Saharan desiccation. The absence of these Canarian lineages nowadays in Africa suggests important demographic movements in the western area of this Continent.
—Maca-Meyer 2003
U6a: it is the most widespread (from Canary Islands and Iberian Peninsula to Syria, Ethiopia and Kenya) and has highest diversity in Eastern Africa. Estimated age: 24-27,500 BP. It has one major subclade:
U6a1: with similar distribution to U6a. Estimated age: 15-20,000 BP. I was also found in Brittany (France) at 4.5% frequency (1/22)[18]
U6b: shows a more patched and western distribution. In the Iberian peninsula U6b is more frequent in the North (while U6a is in the South). It has also been found in low amounts in Morocco, Algeria, Senegal and Nigeria. Estimated age: 8,500-24,500 BP. It has one subclade:
U6b1: found only in the Canary Islands and in the Iberian peninsula. Estimated age: c. 6000 BP.
U6c: only found in Morocco and Canary Islands. Estimated age: 6,000-17,500 BP.
U6a and U6b share a common basal mutation (16219) that is not present in U6c.”
http://en.wikipedia.org/wiki/Haplogroup_U_%28mtDNA%29
What do you think about that?
the Skeptic
Let me state this very clearly – the original ancestors of all humans, INCLUDING Blacks, had dark hair, skin, and eyes – they were MOT classified as any ethnic group – according to the “Out of Africa” theory – blacks wer the first to diversify – taking the most diverse genes with them – Idaltu probably could be considered the beginning of the Black ethnic group – others diversified with different sets of genes forming what lead to other ethnic groups – Cro Magnon in Europe could be considered the beginning of the White, and probably, remainder of the ethnic groups – their is about a 40,000 – to as much as a 100,000 year gap that is unexplained as to how European Cro Magnon got to, or, started in, Europe. There is NO European Cro Maganon in Africa – Idaltu is a different version. NO Neanderthals have been found in Africa,but EVERYWHERE else,to date – AGAIN a hole in the theory along with the older Asian haplomarker M, than any found in Africa.
CN
I addressed my mistake of using Black=’African’=”Negroid”. Clearly, the Euopean derives from The Veddoid after splitting from Pro-Capoid (Khoi-San)mt-DNA L !
http://www.academon.com/Analytical-Essay-A-Prehistoric-Ethnic-Group/106001
From the Paper:
“Although, other groups certainly coexisted on the planet with the white AMH people, none of the remains found have had the DNA confirmations to a white race as the Cro-Magnon. The Homo erectus, Homo ergaster, and the Neanderthals are all considerably different from each other and from the AMH, but could have possibly mixed at various points in time to cause the range in ethnic groups of today. Another belief of how the white AMH people may have developed into other ethnicities without mixing is through the gradual gain in skills and knowledge that led up to settlements in ancient Babylon, possibly during the Neolithic Age.”
AMH – Atlantic Modal Haplotype !
“…the white AMH people, NONE of the REMAINS found have had the DNA confirmations to a WHITE race as the Cro-Magnon.” Note: Hg R is Eurasian/Veddoid not European/white:
“Y-haplogroup R is found throughout all continents, but is fairly common throughout Europe, South Asia and Central Asia. In these regions the distribution is markedly different for the two major subclades R1a and R1b.
It is important in Native Americans and it also occurs in Caucasus, Near East, West China, Siberia and some parts of Africa.
Small frequencies are found in Malaysia, Indonesia, Philippines, Korea and Indigenous Australians.
[Manfred Kayser et al 2002-03, Reduced Y-Chromosome, but Not Mitochondrial DNA, Diversity in Human Populations from West New Guinea]
If you look again, you will see the order is wrong…
Malaysia, Indonesia, Philippines, Korea and Indigenous Australians, since Indigenous Australians are the Veddoid and are first in the OoA Theory.
In other words, what passes as Cro-Magnon is the Veddoid R-M343, R-M207, R-M173, etc in order to mesh with the 28,000 YBP so-called European Cro-Magnon.
The Veddoid phenotype would have been closest to the African R1b even as a back-migration from Asia! This is further circumstantial evidence that the Veddoid/Aboriginal Australian Cro-Magnon was the ancestor of modern Europeans but was a split from its Eurasian genotype-phenotype.
So, I stand corrected:
My use of Black-African is apparently corrected as Y-DNA CT-M168(ancestral)East African Veddoid Phenotype from which all non-African hgs derived.
This reaffirms this…
“Although, other groups certainly coexisted on the planet with the white AMH people, none of the remains found have had the DNA confirmations to a white race as the Cro-Magnon.
As the Cro-Magnon was Phenotypicaly Veddoid, South Asian R-M207, etc., until we get to the so-named R-M269 dominant in ‘European’ populations.
Veddoid: The Phenotype template for Europeans and East Africans with allele differentiation for pigment, hair and eye color!
Like I said, the light goes on, however, when R1* returns in the back-migration to Central Africa the genotype-phenotype is Black-African NOT VEDDOID-Caucasian or European-Caucasian.
the Skeptic
Whether you realize it or NOT – you have presented evidence of what I have said all along all humans are descended from a common ancestor , NOT from Blacks – your problem comes because you keep equating Africans with Blacks – NONE of the information you presented, does this – ancient humanoids according to the “Out of Africa” theory LIVED IN AFRICA – they are AFRICANS – NOT Black Africans. Blacks being the first to diversify have the most common genes, the further out from Africa, the fewer genes in common with the first human ancestor. Like I said before – there are some haplomarkers shared by all ethnic groups, but there are also haplomarkers limited to certain ethnic groups – this applies to subclades to – this is WHY DNA can determine ethnicity. If you examine forensic drawings of the two different Cro Magnons – Idaltu – the African one, could be classified as Black, but is not – the closest living example, as I mentioned before, is the Black Masai tribe of Africa. Cro Magnon, the European one, could be classified as White, but is not – the closest living examples being the Finns, or, Danes.
Lol,he was some damned honkey!?
Kamal Gazzah….FYI
CONDITION OF SKELETON AND PREVIOUS STUDIES ON KING TUT:
“Despite the difference in opinion, both scientists agree on one fact. The mummy is currently in bad condition, and was handled roughly in the past. In 1925, a year after its discovery, a team of professors including Howard Carter tried to examine the mummy. On finding it stuck to the sarcophagus, they put it out in the sun to melt the materials keeping it together. They later used hot knifes to pry the golden mask off the face, the golden bracelets off the arms and toes. As a result, the head was separated from the body, the pelvis from the midriff and so were the arms, hands, legs and feet.”
“In 1968 AND 1978, the mummy was first analyzed and x-rayed by R.G. Harrison from Liverpool University, and second by James Harries from the University of Michigan, Ann Arbor. According to Saleh, the results of the studies were never published in their complete form anywhere, so why do another study? “Enough handling of an already suffering mummy! Why not seek the studies that took place in the past? We must first know what their discoveries were so as to know what to look for,” he argues.”
Take a look at his skeleton and try to put him back together like Humpty-Dumpty!
http://news.nationalgeographic.com/news/bigphotos/39326459.html
“They later used hot knifes to pry the golden mask off the face, the golden bracelets off the arms and toes. As a result, the head was separated from the body, the pelvis from the midriff and so were the arms, hands, legs and feet.”
For that matter how do we know the bone-necrotic disease did not make his bones more porous – open to the falling blood of someone cutting into the bones of his extremities, legs, femure, etc., thereby replacing ancient DNA with modern (but old from 1925) DNA?
“Necrosis typically begins with cell swelling, chromatin digestion, and disruption of the plasma membrane and organelle membranes. Late necrosis is characterized by extensive DNA hydrolysis, vacuolation of the endoplasmic reticulum, organelle breakdown, and cell lysis. The release of intracellular content after plasma membrane rupture is the cause of inflammation in necrosis.”
ADDITIONAL TESTING RE: SICKLE CELL:
“A vascular necrosis of the femoral head is the death of bone that results in the collapse of the architectural bony structure, leading to pain and loss of joint function. Secondary avascular necrosis of the femoral head has a definite cause with an interruption of the vascular blood supply due to local trauma or nontraumatic systemic conditions. All other cases of avascular necrosis of the femoral head are classified into idiopathic primary avascular necrosis of the femoral head. Nontraumatic avascular necrosis of the femoral head is caused by long-term extensive corticosteroid administration, hemoglobinopathies (eg, sickle cell anemia), fat emboli, and alcoholism. (1),(2)”
(1) Assouline-Dayan Y, Chang C, Greenspan A, Shoenfeld Y, Gershwin ME. Pathogenesis and natural history of osteonecrosis. Semin Arthritis Rheum. 2002; 32(2):94-124.
(2) Lafforgue P. Pathophysiology and natural history of avascular necrosis of bone [published online ahead of print August 8, 2006]. Joint Bone Spine. 2006; 73(5):500-507.
If this happens with sickle cell anemia what about Kholer’s disease?
http://www.associatedcontent.com/article/2708235/rare_kohlers_disease_affected_bones_pg2.html?cat=70
“Kohler’s disease seldom lasts more than two years. Bone fragments heal and harden back into solid bone. Beyond rest, over-the-counter medications to relieve discomfort, and avoiding excessive weight-bearing activities, most children require no treatment. The majority of affected children have no long-term consequences.”
Only affects the left foot typically and is NOT LIFE THREATENING!
When was the earlier mt-DNA testing done?
“Sadly the study has failed to answer these questions which brings me to the second thing that puzzles me: no mention is being made of the earlier DNA sampling of key New Kingdom mummies. As I reported in September 2009, Prof Scott Woodward harvested DNA from 27 New Kingdom mummies, 7 successfully. He also successfully recovered mitochondrial DNA from the elder KV62 foetus, although the sequencing has never been published. There are also reports that he successfully extracted DNA from Yuya, the father of Queen Tiye. Nothing I have read suggests that any attempt has been made to integrate these earlier DNA sequences. Undoubtedly methodologies have moved on since the 1990s, but nonetheless I worry whether the politics of “an all Egyptian investigation” may have come before maximising the scientific value of the DNA which has been gathered from New Kingdom mummies. That is particularly true of the foetuses from KV62 because, as I have reported previously, they have allegedly not been well kept and have deteriorated over the past decade making DNA extraction more difficult.”
Posted by Kate Phizackerley on Friday, February 19, 2010
http://www.kv64.info/2010/02/critique-of-tutankhamun-dna-analysis.html
CN
WHAT?!
“Homo sapiens idaltu is an extinct subspecies of Homo sapiens that lived almost 160,000 years ago in Pleistocene Africa.[1] Idaltu is from the Saho-Afar word meaning “elder or first born”.[1]”
BRAIN CAPACITY: “The fossilized remains of H. s. idaltu were discovered at Herto Bouri in the Middle Awash site of Ethiopia’s Afar Triangle in 1997 by Tim White, but were first unveiled in 2003.[1] Herto Bouri is a region of Ethiopia under volcanic layers. By using radioisotope dating, the layers date between 154,000 and 160,000 years old. Three well preserved crania are accounted for, the best preserved being from an adult male (BOU-VP-16/1) having a brain capacity of 1,450 cm3 (88 cu in).”
Can you say homeland Pw-n-Netjer,(Pu-An-Netjer) Land of Punt, North Ethiopia, Massawa, Eritrea?
NOT CRO-MAGNON OF EUROPE:
“These fossils differ from those of chronologically later forms of early H. sapiens such as Cro-Magnon found in Europe and other parts of the world in that their MORPHOLOGY HAS MANY ARCHAIC FEATURES NOT TYPICAL OF HOMO SAPIENS (although modern human skulls do differ across the globe).[1]”
“Despite the archaic features, these specimens were argued to represent the direct ancestors of modern Homo sapiens sapiens which, according to the “recent African origin (RAO)” or “out of Africa” theory, developed shortly after this period [mt-DNA L] (KHOI-SAN mitochondrial DIVERGENCE dated not later than 110,000 BCE) in Eastern Africa. “The many morphological features shared by the Herto crania and AMHS, to the exclusion of penecontemporanous Neanderthals, provide additional fossil data excluding Neanderthals from a significant contribution to the ancestry of modern humans.”[1]
KHOI-SAN = CAPOID = Idaltu (First Born)
If, anything the structure would be something like Idaltu-Capoid, Idaltu-Veddoid and lastly, Idaltu-Europoid (The postulated subraces vary depending on the author, including but not limited to Nordic, Mediterranean, Alpine, Dinaric, East Baltic, Arabid, Turanid, Iranid and Armenoid subraces.)
“An exact description was made, by its discoverers, of Homo sapiens idaltu.
“On the limited available evidence, a subspecies of Homo sapiens distinguished from HOLOCENE anatomically modern humans (Homo sapiens sapiens) by greater craniofacial robusticity, greater anterior–posterior cranial length, and large glenoid-to-occlusal plane distance. Homo sapiens idaltu is distinguished from the holotype of Homo rhodesiensis (Woodward, 1921) by a larger cranial capacity, a more vertical frontal with smaller face, and more marked midfacial topography (for example, canine fossa). We consider the holotypes of H. helmei and H. njarasensis too fragmentary for appropriate comparisons.”[1]
LIST OF FOSSIL SITES
http://en.wikipedia.org/wiki/List_of_fossil_sites
LIST OF HUMAN EVOLUTION FOSSILS:
“Most of the fossils shown are NOT considered DIRECT ANCESTORS to Homo sapiens but are closely related to direct ancestors and are therefore important to the study of the lineage.”
http://en.wikipedia.org/wiki/List_of_human_evolution_fossils
ARCHAIC HOMO SAPIENS:
http://en.wikipedia.org/wiki/Archaic_Homo_sapiens
CN
If you are going to challenge me then at least provide the accurate information and not YOUR INTERPRETATION.
LAND OF PUNT / CHAMPOLLION (Transcriber of the Rosetta Stone):
“Just a few years later, in 1839, Champollion states in his work “Egypte Ancienne” that the Egyptians and Nubians are represented in the same manner in tomb paintings and reliefs and that “The first tribes that inhabited Egypt, that is, the Nile Valley between the Syene cataract and the sea, came from Abyssinia to Sennar. In the Copts of Egypt, we do not find any of the characteristic features of the Ancient Egyptian population. The Copts are the result of crossbreeding with all the nations that successfully dominated Egypt. It is wrong to seek in them the principal features of the old race.”[12]
http://en.wikipedia.org/wiki/Ancient_Egyptian_race_controversy
1820 drawing of a Book of Gates fresco of the tomb of Seti I, depicting (from left): Libyan, Nubian, Asiatic, Egyptians.
ANTHROPOLOGY:
“Several anthropologists who study the biological relationships of the Ancient Egyptian population call for a recognition of Africa’s genetic diversity when considering the racial identity of the Ancient Egyptians.[44]”
KING TUT’S AFRICAN ANCESTRY:
“Ahmed Saleh, the former archaeological inspector for the Supreme Council of antiquities stated that the procedures used in the facial re-creation made Tut look Caucasian, “disrespecting the nation’s African roots.”[54]”
We can disagree over which source we chose to accept but it remains intellectually dishonest to provide only one side and then to alter the source with your OPINION! That does not make you righteously right because you you have been convinced of your own opinion. It does however, make you someone who cannot measure the informational chronological genetic, anthropological, and historical migration patterns of source materials.
You cannot put any ‘looks like’ Veddoid description that suits you when such a phenotype has already been confirmed:
“The Australoid race is a broad racial classification. The concept originated with a typological method of racial classification.[1][2][3] They were described as having dark skin with wavy hair, in the case of Aboriginal Australians, or hair ranging from straight to kinky in the case of Papuan, Melanesian and Negrito groups.”
RELIABLE DNA FOR KING TUT?
“In 1925, a year after its discovery, a team of
professors including Howard Carter tried to
examine the mummy. On finding it stuck to the
sarcophagus, they put it out in the sun to melt
the materials keeping it together. They later
used hot knifes to pry the golden mask off the
face, the golden bracelets off the arms and toes.
As a result, the head was separated from the
body, the pelvis from the midriff and so were
the arms, hands, legs and feet.”
On what planet do you usually reside, CN? There is more than enough evidence to strongly suggest that DNA testing on King Tut is not reliable until an independent laboratory conducts testing.
I thought this blog was a search for truth, but again the light goes on and it is clear that you are more like the fingers that type the opinions of Dr. Hawass on this site while you simultaneously pretend to disagree with him.
Beyond Thunderdome: CN actually took your sarcasm seriously…
Crips…crypts…ROFLMAO.
Beyond Thunderdome
Neither – they don’t accept Whites(Semitic), and, those two gangs, did NOT exist at that time, and, do NOT exist in the Middle east.
Was he a Crip or a Blood?
Kamel Gazzah
http://www.bioforensics.com/downloads/index.html
DNA term Linkage:
“The association of alleles at two or more loci due to their residing on a single chromosome or their abundance in a particular ethnic group, that causes them to appear together at a higher than expected frequency.”
As I stated, if it is all about higher frequencies in a particular population, than King Tut
D13S317 = 12, 10 : King Tut, Younger Lady, and KV55
Google Search :Population data on the thirteen CODIS core short tandem
Ethnic Allele Estimation – Data Base Locus Pair African American : D13S317/D3S1358. Compare:
D13S317=12
African American Population 48.32
Caucasian – 30.86
U.S Hispanic – 21.67
D13S317=10
African American – 5.28
Caucasian – 5.102
U.S Hispanic – 21.65
What ethnicity was King Tut?
Search Population Frequencies:
http://www.allelefrequencies.net/pop6002a.asp
Understanding Allele frequencies:
http://www.uic.edu/classes/bms/bms655/lesson13.html
http://www.dartmouth.edu/~chance/teaching_aids/books_articles/DNAtyping/node1.html
I hope all of this helps to show that not everything is as simple as some African-centrists and Euro-centrists would like it to be about ancestry.
the Skeptic
I don’t know about you – seems the light bulb flickers in your head, than goes out – what I said is, there are two forms of Cro Magnon – Idaltu in Africa – NOT classified as, but appears Black, and, Cro Magnon, in Europe, NOT classified as, but appears White. Ancient Egyptians were Arab Semitic White(generally, olive colored(golden brown skin – MUCH darker in sunny places as Egypt), dark hair, and eyes – 80% – 20% look like the gamut of Northern Europeans – UNMIXED. Southern Europeans look like, and, are, often confused as Arabs, and, vice versa. Generally. the people of India, and, Pakistan are classified as White – many now see admixtures aof Black, White, and, Oriental Asian in them the Veddoid – you speak of. The Black Nubians and Ancient egyptians(Arab Semitic White) did in fact produce mixes – – I have REPEATEDLY stated this – 65% of Egypt’s Ancient, and, Modern poplulation, is Arab Semitic White, 30% is Black, Black/Arab mixes, 5% all others.
Kamal Gazzah
you are a late comer – R1b1 ORIGINATES in the Middle East. There are three subclades – possibly a new fourth may have been found – two occur in Blacks, one in Caucasians – that is the one King Tut has – NO, it is NOT Western European. It is the one that ORIGINATES in the Middle East – apparently, Europeans have higher amounts of it now, but its origins are in the Middle east – no evidence of mixing occurring – definitely possible. Incidentally, Carol Thatcher’s, daughter of Margaret Thatcher, DNA shows she is 24% ARAB : http://www.margaretthatcher.org/document/110880. Compare Michael Jackson to King Tut in his midway plastic surgery change to becoming artificially Caucasian looking : http://naturescorner.wordpress.com/2008/12/13/where-the-ancient-egyptians-black-or-white/
R1b Haplogroup is not specific to Western Europe, it is also found in North Africa, Middle East (including Egypt) and sub Sahara (Chad, Soudan, Cameroun), India etc.
So TUT Haplogroup is not specifically Western European
One more thing it is not proved that the Haplotype showed by the Video is TuT’s one and could be for demonstartion only.
Wow, that means the Caucasian is the Veddoid phenotype Cro-Magnon, while ‘Europeans’ are white. Based only this understanding the ancient Egyptians were Caucasian but phenotypically a middle type between Sub-Saharan Africans and white Europeans.
Lightbulb moment. Maybe I am a little slow but will concede you were right about an indiscriminate use of Black=African.
CN
…okay…here is something relevant to DNA studies..also shows a map of Pre-historic Movements of Ethnic Groups….
http://dnaconsultants.com/_blog/DNA_Consultants_Blog/post/Emerging_Prehistory_of_Ethnic_Groups/
“From what we can see, putting on diachronistic lenses, the mutation rates for the DYS values on what are commonly called CODIS markers or the DNA profile for individuals are very small. The values appear to have been set from the beginning of mankind and to have mutated little in the past 100,000 years.
If this is true — and it cannot be a very big “if” or we would have more diversity between populations than what is known — the oldest markers are Sub-Saharan African and the newest European.
Statistical divides bear out this reading of the human genetic record, as shown now in our updated map included with the DNA Fingerprint Plus.”
Also this one…Peking Man, a Homo erectus fossil unearthed outside Beijing.
Read more: http://www.time.com/time/world/article/0,8599,2054949,00.html#ixzz1Spcpu3hr
“These days, the claims are getting a rough shaking from hard science–as is the notion that the Chinese are a unique, indigenous race. The Chinese Human Genome Diversity Project, a collaboration among 12 researchers from seven institutions, scrutinized DNA samples from 28 of China’s 56 ethnic groups and then compared the samples with genetic material from other Asian and non-Asian groups. Their verdict: Chinese–like the rest of humanity–evolved in Africa. They migrated eastward along the Indian Ocean and made their way to China via Southeast Asia.
… It is now safe to conclude, reported the researchers, that modern humans originating in Africa constitute the majority of the current gene pool in Asian populations.”
Read more: http://www.time.com/time/world/article/0,8599,2054949,00.html#ixzz1SpcVSVLW
And this one…
http://www.academon.com/Analytical-Essay-A-Prehistoric-Ethnic-Group/106001
From the Paper:
“Although, other groups certainly coexisted on the planet with the white AMH people, none of the remains found have had the DNA confirmations to a white race as the Cro-Magnon. The Homo erectus, Homo ergaster, and the Neanderthals are all considerably different from each other and from the AMH, but could have possibly mixed at various points in time to cause the range in ethnic groups of today. Another belief of how the white AMH people may have developed into other ethnicities without mixing is through the gradual gain in skills and knowledge that led up to settlements in ancient Babylon, possibly during the Neolithic Age.”
AMH – Atlantic Modal Haplotype !
“…the white AMH people, none of the remains found have had the DNA confirmations to a white race as the Cro-Magnon.” Note: Hg R is Eurasian not European:
“Y-haplogroup R is found throughout all continents, but is fairly common throughout Europe, South Asia and Central Asia. In these regions the distribution is markedly different for the two major subclades R1a and R1b.
It is important in Native Americans and it also occurs in Caucasus, Near East, West China, Siberia and some parts of Africa.
Small frequencies are found in Malaysia, Indonesia, Philippines, Korea and Indigenous Australians.
[Manfred Kayser et al 2002-03, Reduced Y-Chromosome, but Not Mitochondrial DNA, Diversity in Human Populations from West New Guinea]
If you look again, you will see the order is wrong…
Malaysia, Indonesia, Philippines, Korea and Indigenous Australians, since Indigenous Australians are the Veddoid and are first in the OoA Theory.
In other words, what passes as Cro-Magnon is the Veddoid R-M343, R-M207, R-M173, etc in order to mesh with the 28,000 YBP so-called European Cro-Magnon.
The Veddoid phenotype would have been closest to the African R1b even as a back-migration from Asia! This is further circumstantial evidence that the Veddoid/Aboriginal Australian Cro-Magnon was the ancestor of modern Europeans but was a split from its Eurasian genotype-phenotype.
So, I stand corrected:
My use of Black-African is apparently corrected as Y-DNA CT-M168(ancestral)East African Veddoid Phenotype from which all non-African hgs derived.
This reaffirms this…
“Although, other groups certainly coexisted on the planet with the white AMH people, none of the remains found have had the DNA confirmations to a white race as the Cro-Magnon.
As the Cro-Magnon was Phenotypicaly Veddoid, South Asian R-M207, etc., until we get to the so-named R-M269 dominant in ‘European’ populations.
Veddoid: The Phenotype template for Europeans and East Africans with allele differentiation for pigment, hair and eye color!
Veddoid: A member of an ancient race of southern and southeastern Asia and northern Australia, characterized by dark brown skin, slim build, and wavy hair.
“In the out of Africa theory, the ancestors of the Australoids, the Proto-Australoids are thought to have been the first branch off from the Proto-Capoids to migrate from Africa about 60,000 BCE, migrating along the now submerged continental shelf of the northern shore of the Indian Ocean and reaching Australia about 50,000 BCE.”
Capoids = San of South East Africa, obviously already having the lighter complexion genotype-phenotype before migrating OoA.
How is that for not being Multi-Regionalism?
the skeptic
Honestly – the best I can do, is give you a Google search – much indicates different ethnic groups among prehistoric peoples – most indicate Neanderthals as one such example, non comes righ out and says so – like Idaltu – African Cro Magnon, and, European Cro Magnon we discussed earlier: http://www.google.com/search?q=When+did+different+ethnic+groups+form%3F&ie=utf-8&oe=utf-8&aq=t&rls=org.mozilla:en-US:official&client=firefox-a#sclient=psy&hl=en&client=firefox-a&hs=FnO&rls=org.mozilla:en-US%3Aofficial&source=hp&q=Prehistoric+ethnic+groups%3F&aq=f&aqi=&aql=&oq=&pbx=1&bav=on.2,or.r_gc.r_pw.&fp=aeb16183bfd07c66&biw=998&bih=621
Beyond Thunderdome
Please, explain your view more clearly.
He looks to be at peace,though probably never saw it in his own time.
These other Mummies of the cocaine lands always look to know no peace,it’s like arrgh! no more coca for us!
CN
Please site your source for this:
“… – just different skeletal proportions setting off the ethnic groups.”
Is this your way of pushing the Multi-Regional Theory?
CN
Actually, you should change that from “..in general, the further away from Africa, the fewer of the dark skin color genes…”
to
the further one moves away from the equator…
“Nature has selected for people with darker skin in tropical latitudes, especially in nonforested regions, where ultraviole radiation from the sun is usually the most intense. Melanin acts as a protective biological shield against ultraviolet radiation. By doing this, it helps to prevent sunburn damage that could result in DNA changes and, subsequently, melanoma a cancer of the skin. Melanoma is a serious threat to life. In the United States, approximately 54,000 people get this aggressive type of cancer every year and nearly 8,000 of them die from it. Those at highest risk are European Americans. They have a 10 times higher risk than African Americans.”
And, African Americans are ‘Black’ – not the color…you know exactly what I mean.
You fella are obsessing over the dead.
The skeptic – I will cede to you on that one – yes – darker skin was first -the Europeans and Asians diverged of the ancestral lines, then developed similar lighter skin for different reasons (convergence). Incidentally, darker skin does NOT equate with Black – Blacks being the first to diverge, carried the most genes for dark skin color – in general, the further away from Africa, the fewer of the dark skin color genes, so the theory goes – whether do to environment, hereditary, or both, is still open to speculation, as is even the “Out of Africa” theory. genes for light sklin, hair, and eyes is a mutation that occurred about 13,000 – 15, 000 years ago – all ethnic groups had dark hair, skin, and eyes until then – just different skeletal proportions setting off the ethnic groups.
CN…you wrote..
”Our data strongly support independent genetic origins for the light skin phenotype in Europeans and East Asians arising after the divergence of modern European and East Asian populations.”
Then you are saying that the LIGHT SKIN skin of Europeans and East Asians arose AFTER the DIVERGENCE of modern EUROPEAN and East ASIAN populations MEANS the ancestral allele for color was that of the darker complexioned one….from Africa into Southeast Asia…so I do not understand why you say I am wrong? Black skin came first and then lighter complexions after the Eurasian population diverged to become European and Asian.
Sometimes I think you just like to misquote and mis-interpret what I post for the heck of it and that is pretty weak dude.
You should really think about what you read before you alter it to suit an opinion. The point was that the first skin color/complexion was dark due to adaptation to UV light. As the OoA happened the need for lighter complexions became adaptive to northern climates where UV rays were obscured in those climates.
the Skeptic – your reasoning is wrong – the last sentence taken from the “Conclusion) of the actual study explains WHY: Quote,”Our data strongly support independent genetic origins for the light skin phenotype in Europeans and East Asians arising after the divergence of modern European and East Asian populations.”: http://mbe.oxfordjournals.org/content/24/3/710.full
The study was to determine if skin color is determined by divergence(ancestral lineage), or, convergence(common denominator producing similar changes in two populations – Europeans, and, East Asians. The study does vaguely refer to admixture of Native indians playing a factor to. The study seems to indicate, outside of Africa,it was convergence, NOT divergence – meaning possible factors other than hereditary, played a factor – i.e. – mate selection for lighter skin tones (not quite worded like this – that is the idea though)
CN…update on genotype-phenotype re: Skin Color
Ancestral Phenotype:
“The ADTB3A gene also shows a strong and focused signature of positive selection in Africans…
Many hypotheses predict that natural selection will eliminate genetic variants associated with lighter skin in the regions of high UVR as a protection against photo damage (e.g., sunburn, melanoma, and basal and squamous cell carcinomas) (Blum 1961; Kollias et al. 1991) and folic acid photo degradation (Branda and Eaton 1978; Jablonski and Chaplin 2000). The photo protective properties of a highly melanized skin and the recent African origin of modern humans suggest that the ancestral phenotype is one of the relatively dark skin (Jablonski and Chaplin 2000; Rogers et al. 2004). If dark skin is the ancestral phenotype, then we may assume that the first migrants out of Africa were relatively darkly pigmented…”
Skin color, i.e., phenotype mutations:
“In general, the DERIVED allele (associated with lighter pigmentation) is most common in Europeans and East Asians, and the ANCESTRAL allele predominates in sub-Saharan Africa and Island Melanesia.”
I hate to be nitpicky but shouldn’t this sentence been written:
“In general, the ANCESTRAL allele predominates in sub-Saharan Africa and Island Melanesia, and the DERIVED allele (associated with lighter pigmentation) is most common in Europeans and East Asians.”
Why? Because ancestral is first but for some reason writers put the European derived genotypes before the ancestral genotypes. It is intellectually dishonest to say the least, particularly if the average person does not understand that ancestral is at the top of the tree of life and derived are the branches below (downstream, so to speak).
Africans – Ancestral
Europeans – Derived
(Continue…)
The Skeptic
Do you READ/Comprehend what you copy/paste? – The “Land of Punt is SEPARATE from Ancient Egypt – the Ancient Egyptians traded with them – the Puntians are NOT the Ancient Egyptians – the Land of Punt is part of what is NOW Ethiopia, Eritrea, and, some say Somalia, and Sudan , NOT Ancient Egypt – AGAIN, you are grasping for straws. The people of Chad received their R1b by back flow from Western Asia(part of Asia that is the Middle(formerly Near) East as in Arab Semitic White:
http://en.wikipedia.org/wiki/Haplogroup_R1b_%28Y-DNA%29
http://www.egyptsearch.com/forums/ultimatebb.cgi?ubb=get_topic;f=8;t=007225
CN
There you go again…not interpreting what was given….The Land of Punt is proven to be in Eritrea, which is the ancestral homeland of the ancient so-called Egyptians.
Tsk, tsk…
the Skeptic
Guess you were attempting to indicate the Land of Punt was Egypt, and, it was Black – YOUR own link indicates this is NOT so – it was located OUTSIDE of Egypt- MORE,and MORE RECENT evidence of this:http://www.twcenter.net/forums/showthread.php?t=352871
Land of Punt
http://wysinger.homestead.com/punt.html
CN – BELATED UPDATE
“Baboon Mummy Tests Reveal Ethiopia and Eritrea as Ancient Egyptians’ ‘Land of Punt’
Submitted by owenjarus on Fri, 04/23/2010 – 15:02
Mummified baboon at the British museum
Copyright the Trustees of the British Museum. A baboon mummy from the Temple of Khons at Karnak in Thebes – it dates to the New Kingdom. It’s one of three that the British Museum has. The scientists were given permission to test the hairs from two of them.
Heritage Key reported recently that mummified baboons in the British Museum could reveal the location of the land of Punt – a place to which pharaohs organized trading expeditions. To the Egyptians, Punt was a place of fragrances, giraffes, electrum and other exotic goods. It was sometimes referred to as Ta-Netjer – ‘God’s land’ – a huge compliment given that the Ancient Egyptians tended to view outside cultures with disdain.”
Read entire article here:
http://heritage-key.com/blogs/owenjarus/baboon-mummy-tests-reveal-ethiopia-and-eritrea-ancient-egyptians-land-punt
“Although Egyptians record voyaging to it until the end of the New Kingdom, 3,000 years ago, scholars do not know where Punt was. Ancient texts offer only vague allusions to its location and no ‘Puntite’ civilization has yet been discovered. Somalia, Ethiopia, Yemen and even Mozambique have all been offered as possible locations.
Thanks to some cutting edge science, the search for Punt appears to be coming to an end. New research, to be presented at an Egyptology conference today, provides proof that it was located in Eritrea/East Ethiopia.
The researchers compared the oxygen isotope values in the ancient baboons to those found in their modern day brethren.”
…
“All of our specimans in Eritrea and a certain number of our specimens from Ethiopia – that are basically due west from Eritrea – those are good matches,” said Professor Dominy.
“We think Punt is a sort of circumscribed region that includes eastern Ethiopia and all of Eritrea.”
Somalia, Yemen and Mozambique do not match.”
SEE : “Photo by Hansueli Krapf. A shot of Eritrea, at a point between Asmara and Massawa. New research has identified this as “Punt” – God’s land. CC Attribution share-alike 3.0 unported.”
Geographic Coordinates :15°36′33″N 39°26′43″E : Use Google Maps/Google Earth! Zoom out for earth view.
Cultural dis-information:
“It was sometimes referred to as Ta-netjer – ‘God’s land’ – a huge compliment given that the Ancient Egyptians tended to view outside cultures with disdain.”
One could only come to this conclusion by assuming the ancient Egyptians were not Eritreans/Ethiopians.
Queen Ati/Eti of Punt :
http://en.wikipedia.org/wiki/Land_of_Punt
Ancestral Home
Ha-Tje-Sua /Hatasu / Het-Shep-Sua Ta-Netjer :
http://digital.library.upenn.edu/women/edwards/pharaohs/pharaohs-8.html
Deir El Bahari / Deir Ba-Hari-Ta-Anu (Haritans):
http://www.historykb.com/Uwe/Forum.aspx/general-history/4266/Was-the-Queen-of-PUNT-seen-in-Hatshepsutes-Temple-relief
“According to the temple reliefs, the Land of Punt was ruled at that time by King Parahu and Queen Ati.[11] This well illustrated expedition of Hatshepsut occurred in Year 9 of the female pharaoh’s reign with the blessing of the god Amun:
“ Said by Amen, the Lord of the Thrones of the Two Land: ‘Come,come in peace my daughter, the graceful, who art in my heart, King Maatkare [ie. Hatshepsut]…I will give thee Punt, the whole of it…I will lead your soldiers by land and by water, on mysterious shores, which join the harbours of incense…They will take incense as much as they like. They will load their ships to the satisfaction of their hearts with trees of green [ie. fresh] incense, and all the good things of the land.'”
The more times passes the more truth comes out to reveal the gaps in our knowledge!
Language correction: Parahu to (Amon-Mut-Ta-Pa-Ras-Sahu) texts as Mon-o-mutapa!
Can also be written Amon-Mut-Ta-Pa-Sahu Ras (Sahure/Sahura)!
http://en.wikipedia.org/wiki/Sahure
CN…
http://www.isogg.org/tree/ISOGG_HapgrpR.html
Y-DNA Haplogroup R and its Subclades – 2011 UPDATED!
“Paragroup is a term used in population genetics to describe lineages within a haplogroup that are not defined by any additional unique markers. In human Y-chromosome DNA haplogroups, paragroups are typically represented by an asterisk (*) placed after the main haplogroup[1]. The term “paragroup” is a portmanteau of the terms paraphyletic haplogroup indicating that paragroups form paraphyletic subclades.[1] Apart from the mutations that define the parent haplogroup, paragroups may not possess any additional unique markers. Alternatively paragroups may possess unique markers that have not been discovered. If a unique marker is discovered within a paragroup, the specific lineage is given a unique name and is moved out of the paragroup to form an independent subclade.”
http://en.wikipedia.org/wiki/Paragroup
“…example is a member of the Y-DNA haplogroup R (defined by marker M207) may belong to the sub-haplogroup R1 (defined by marker M173) or R2 (defined by marker M124). Individuals with neither of these mutations would be categorised as belonging to group R*.”
Independent Sub-Clade – That is a separate branch on the R1b1 Tree.
“Paragroup R1b1* and haplogroup R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.”
…V88 is the definitive Afrikan marker having a coalescence time estimated at 9200–5600 kya.
http://groups.yahoo.com/group/scotch-irish/message/54001
2010 – R-V88 (R1b1a). Sub-Saharan African R1b.
2011 – R-V88 (R1b1c). Sub-Saharan African R1b.
2011 R1b1c was R1b1a 2010.
King Tut – Haplogroup is R1b1b2. R1b1b2 is the old R1b1c which is M269. The old R1b1c-M269 is now assigned to R1b1c-R-V88! Waiting for the study that shows King Tut was not the updated R1b1a2-M269…..
“While Western Europe is dominated by the R1b1a2 (R-M269) branch of R1b, the Chadic-speaking area in Africa is dominated by the branch known as R1b1c (R-V88). These represent two very successful “twigs” on a much bigger “family tree”.
http://www.members.cox.net/generalbanks/completecategorieshapr.html
Markers : (R-P297), (R-M269), (R-M335), (R-V88/V88*), etc.
Paragroup R1b1* ; Descendants:
R1b1a (R-P297)
R1b1a2(R-M269)NGeographic Screen Shot – DNA Test
All this means is this marker was on the screen!
R1b1b (R-M335)
R1b1c (R-V88/V88*)- Paragroup – Siwa Berbers 50.6%
R1b1c1-M18
R1b1c2-V35
R1b1c3* Paragroup
R1b1c3a-V7
R1b1c4-V69
Berbers from Siwa Egypt AA/Berber -93/Total 28.0%
R1b1c (R-V88)- 26.9%
R1b1a2 (R-M269)- 1.1%
R1b1c* (R-V88*)- 23.7%
R1b1a4 (R-V69 – 3.2%
2011 Changes in European R1b according to ISOGG Y-DNA Tree :
“Haplogroup R1b1a2-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia. R1b1a2-M269 is estimated to have arisen approximately 8,000 to 4,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in haplogroup R1b1a2a1a1-L11/S127 and most European R1b1a2 belongs to haplogroups R1b1a2a1a1a-S21/U106 or R1b1a2a1a1b-P312/S116.”
“Population studies which test for M269 have become more common in recent years, while in earlier studies men in this haplogroup are only visible in the data by extrapolation of WHAT IS LIKELY. The following gives a summary of most of the studies which specifically tested for M269, showing its distribution in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.”
R-M73 (R1b1a1). Found in Anatolia, Caucasus, Urals, Hazara
“In a study of Jordan it was found that no less than 20 out of all 146 men tested (13.7%), including most notably 20 out of 45 men tested from the Dead Sea area, were POSITIVE for M173 (R1) but NEGATIVE for P25 and M269, mentioned above, as well as the R1a markers SRY10831.2 and M17, so they are either R1b* or R1a*.[15] Hassan et al. (2008) found an equally surprising 14 out of 26 (54%) of Sudanese Fulani who were M173(Positive+)and P25(Negative-).”
Wood et al. reported high frequencies of men who were P25 POSITIVE and M269 NEGATIVE, amongst the same north Cameroon area where Cruciani et al. reported high R-V88 levels. However they also found such cases amongst 3% (1/32) of Fante from Ghana, 9% (1/11) of Bassa from southern Cameroon, 4% (1/24) of Herero from Namibia, 5% (1/22) of Ambo from Namibia, 4% (4/92) of Egyptians, and 4% (1/28) of Tunisians.”
You can keep saying R1b* is white-Semitic-Arab but the studies prove you are %100 percent wrong also the findings of M269 in Afrikans shows no European admixture. We need to know whether King Tut tested POSITIVE M269 or NEGATIVE M269. If negative…well…there goes the speculation…
Thank you for once again giving me the opportunity to get this information on the record again so that folks who really want to know the truth can find information which will help them to decide for themselves based on the law of probabilities of not high frequencies of the European M269 in the present so-called Arab-Egyptian population.
R1b* Paragroup – You Tube – Chad, Cameroon, and probably includes the aforementioned Shuwa Arabs who are not White-Semitic-Arabs who married Black-Afrikan women. That is the media dis-information you failed to recognize in my earlier post.
WATCH VIDEO
http://www.youtube.com/watch?v=0weSbf0TsZ0
NorthAfrican & welder
R1B1 originates in Arab Semitic White Middle east.
CN…
http://www.isogg.org/tree/ISOGG_HapgrpR.html
Y-DNA Haplogroup R and its Subclades – 2011 UPDATED!
“Paragroup is a term used in population genetics to describe lineages within a haplogroup that are not defined by any additional unique markers. In human Y-chromosome DNA haplogroups, paragroups are typically represented by an asterisk (*) placed after the main haplogroup[1]. The term “paragroup” is a portmanteau of the terms paraphyletic haplogroup indicating that paragroups form paraphyletic subclades.[1] Apart from the mutations that define the parent haplogroup, paragroups may not possess any additional unique markers. Alternatively paragroups may possess unique markers that have not been discovered. If a unique marker is discovered within a paragroup, the specific lineage is given a unique name and is moved out of the paragroup to form an independent subclade.”
http://en.wikipedia.org/wiki/Paragroup
“…example is a member of the Y-DNA haplogroup R (defined by marker M207) may belong to the sub-haplogroup R1 (defined by marker M173) or R2 (defined by marker M124). Individuals with neither of these mutations would be categorised as belonging to group R*.”
Independent Sub-Clade – That is a separate branch on the R1b1 Tree.
“Paragroup R1b1* and haplogroup R1b1c-V88 are found most frequently in SW Asia and Africa. The African examples are almost entirely within R1b1c and are associated with the spread of Chadic languages.”
…V88 is the definitive Afrikan marker having a coalescence time estimated at 9200–5600 kya.
http://groups.yahoo.com/group/scotch-irish/message/54001
2010 – R-V88 (R1b1a). Sub-Saharan African R1b.
2011 – R-V88 (R1b1c). Sub-Saharan African R1b.
2011 R1b1c was R1b1a 2010.
King Tut – Haplogroup is R1b1b2. R1b1b2 is the old R1b1c which is M269. The old R1b1c-M269 is now assigned to R1b1c-R-V88! Waiting for the study that shows King Tut was not the updated R1b1a2-M269…..
“While Western Europe is dominated by the R1b1a2 (R-M269) branch of R1b, the Chadic-speaking area in Africa is dominated by the branch known as R1b1c (R-V88). These represent two very successful “twigs” on a much bigger “family tree”.
http://www.members.cox.net/generalbanks/completecategorieshapr.html
Markers : (R-P297), (R-M269), (R-M335), (R-V88/V88*), etc.
Paragroup R1b1* ; Descendants:
R1b1a (R-P297)
R1b1a2(R-M269)NGeographic Screen Shot – DNA Test
All this means is this marker was on the screen!
R1b1b (R-M335)
R1b1c (R-V88/V88*)- Paragroup – Siwa Berbers 50.6%
R1b1c1-M18
R1b1c2-V35
R1b1c3* Paragroup
R1b1c3a-V7
R1b1c4-V69
Berbers from Siwa Egypt AA/Berber -93/Total 28.0%
R1b1c (R-V88)- 26.9%
R1b1a2 (R-M269)- 1.1%
R1b1c* (R-V88*)- 23.7%
R1b1a4 (R-V69 – 3.2%
2011 Changes in European R1b according to ISOGG Y-DNA Tree :
“Haplogroup R1b1a2-M269 is observed most frequently in Europe, especially western Europe, but with notable frequency in southwest Asia. R1b1a2-M269 is estimated to have arisen approximately 8,000 to 4,000 years ago in southwest Asia and to have spread into Europe from there. The Atlantic Modal Haplotype, or AMH, is the most common STR haplotype in haplogroup R1b1a2a1a1-L11/S127 and most European R1b1a2 belongs to haplogroups R1b1a2a1a1a-S21/U106 or R1b1a2a1a1b-P312/S116.”
“Population studies which test for M269 have become more common in recent years, while in earlier studies men in this haplogroup are only visible in the data by extrapolation of WHAT IS LIKELY. The following gives a summary of most of the studies which specifically tested for M269, showing its distribution in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.”
R-M73 (R1b1a1). Found in Anatolia, Caucasus, Urals, Hazara
“In a study of Jordan it was found that no less than 20 out of all 146 men tested (13.7%), including most notably 20 out of 45 men tested from the Dead Sea area, were POSITIVE for M173 (R1) but NEGATIVE for P25 and M269, mentioned above, as well as the R1a markers SRY10831.2 and M17, so they are either R1b* or R1a*.[15] Hassan et al. (2008) found an equally surprising 14 out of 26 (54%) of Sudanese Fulani who were M173(Positive+)and P25(Negative-).”
Wood et al. reported high frequencies of men who were P25 POSITIVE and M269 NEGATIVE, amongst the same north Cameroon area where Cruciani et al. reported high R-V88 levels. However they also found such cases amongst 3% (1/32) of Fante from Ghana, 9% (1/11) of Bassa from southern Cameroon, 4% (1/24) of Herero from Namibia, 5% (1/22) of Ambo from Namibia, 4% (4/92) of Egyptians, and 4% (1/28) of Tunisians.”
You can keep saying R1b* is white-Semitic-Arab but the studies prove you are %100 percent wrong also the findings of M269 in Afrikans shows no European admixture. We need to know whether King Tut tested POSITIVE M269 or NEGATIVE M269. If negative…well…there goes the speculation…
Thank you for once again giving me the opportunity to get this information on the record again so that folks who really want to know the truth can find information which will help them to decide for themselves based on the law of probabilities of not high frequencies of the European M269 in the present so-called Arab-Egyptian population.
R1b* Paragroup – You Tube – Chad, Cameroon, and probably includes the aforementioned Shuwa Arabs who are not White-Semitic-Arabs who married Black-Afrikan women. That is the media dis-information you failed to recognize in my earlier post.
WATCH VIDEO
http://www.youtube.com/watch?v=0weSbf0TsZ0
the Skeptic
AGAIN, you are NOT comprehending what you are copying and pasting – Rlb1 originates in the Middle/Near East. Your data shows the Arabs spread from North Africa(part of the Middle/Near East) intermarrying with the Blacks to create the mixed tribes listed- “Semitized” as YOUR article calls it. J*, J1, and J2 is an Semitic White(Arabs and Jews) haplomarker – IT DOES NOT OCCUR IN BLACKS – ONLY in Semitic Whites – in YOUR article”s case – ARAB SEMITIC WHITE – you don’t even understand the article is talking about MIXED peoples -Mulattos – Black/ Arab Semitic Whites – NO SUCH THING as an ethnic Black Arab – ONLY MIXED – Mulatto – Black/Arab Semitic White.
Cos Seven
King Tut has been portrayed with black, brown and with tha.
Really,no,he may have been Pharoah.
Or simply a body the embalmers had knocking about.
Their own family.
Not even in a billionaire year!
Tut was NOT of R1B type.
“The DNA test results were inadvertently revealed on a Discovery Channel TV documentary filmed with Hawass’s permission — but it seems as if the Egyptian failed to spot the giveaway part of the documentary which revealed the test results.”
LMAO. The video was faker than a $3 bill.
Hawass is consertive so
1) why did they shot the documentary in the labo?
2) why did they let the camera filming the screen
3) Since that hawass must had been the first to see what was filmed why didnt he deleted the supposed screenshots?
It was obviously made on PURPOSE. Everything is fake about Zahi Hawass.
Tut being Western europeans ? LMAO Hawass approved the reconstution of Tut looking like a modern copt.
Copts do not even look like Western Europeans.
Super fail.
Tut like all the other pharaohs of the XVIII dynasty were BLACK! they were of fallahin/beja type.
In fact they even claimed to be from Punt which was eritrea/ethiopia.